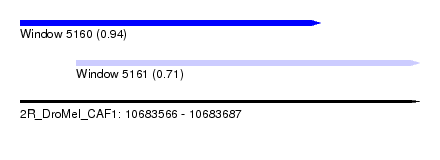
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,683,566 – 10,683,687 |
| Length | 121 |
| Max. P | 0.942124 |

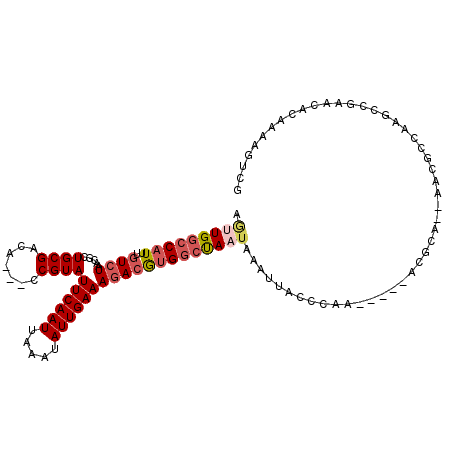
| Location | 10,683,566 – 10,683,657 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 77.54 |
| Mean single sequence MFE | -24.03 |
| Consensus MFE | -17.04 |
| Energy contribution | -17.65 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.942124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10683566 91 + 20766785 GC-----CAGCUG----GAGCAGCAGAGUUGGCCAUUUGUCUAACGCUGCGACA---CCGUAUUCAAUUAAAUAUUGAAAGACGUGGCUAAUAAAUUACCCAA ((-----((..((----..(((((...(((((.(....).))))))))))..))---.(((.((((((.....))))))..)))))))............... ( -24.60) >DroPse_CAF1 23246 94 + 1 GG-----CACUGG-CAGUGGCACUGGCACUGGCCAUUUGUCUGGCCCUGCGACA---CCGUAUUCAAUUAAAUAUUGAAAGACAUGGCCAAUAAAUUACCCAA ((-----((.(((-(((((.(...).)))).))))..))))(((((.((((...---.))))((((((.....))))))......)))))............. ( -27.40) >DroEre_CAF1 20808 96 + 1 GCUGGAGCAGCUG----GAGCAGCAGAGUUGGCCAUUUGUCUAACGCUGCGACA---CCGUAUUCAAUUAAAUAUUGAAAGACGUGGCUAAUAAAUUACCCAA ..(((....((((----...))))...(((((((((..((((.....((((...---.))))((((((.....))))))))))))))))))).......))). ( -27.00) >DroYak_CAF1 21329 93 + 1 GC-----CAGCUGCC--AACCGGCAGAGUUGGCCAUUUGUCUAACGCUGCGACA---CCGUAUUCAAUUAAAUAUUGAAAGACGUGGCUAAUAAAUUACCCAA ..-----...(((((--....))))).(((((((((..((((.....((((...---.))))((((((.....)))))))))))))))))))........... ( -28.70) >DroMoj_CAF1 35329 77 + 1 -------------------GUGGCGACGAUGGCCAUUUAUCUA-UGCUGCGACA---GCGUAUUCAAUUAAAUAUUGAAACACGAGGCUAAUAAAUUACU--- -------------------.((....)).(((((.......((-(((((...))---)))))((((((.....))))))......)))))..........--- ( -17.30) >DroAna_CAF1 20078 87 + 1 GU-----CGUGU----------GCUGUCUUGGCCAUUUGUCUAACGCUGCGACACCACCGUAUUCAAUUAAAUAUUGAAAGACGUGGCUAAUAAAUUACCCA- ..-----.(.((----------(.....((((((((..((((.....((((.......))))((((((.....)))))))))))))))))).....))).).- ( -19.20) >consensus GC_____CAGCUG_____AGCAGCAGAGUUGGCCAUUUGUCUAACGCUGCGACA___CCGUAUUCAAUUAAAUAUUGAAAGACGUGGCUAAUAAAUUACCCAA ...........................(((((((((..((((.....((((.......))))((((((.....)))))))))))))))))))........... (-17.04 = -17.65 + 0.61)

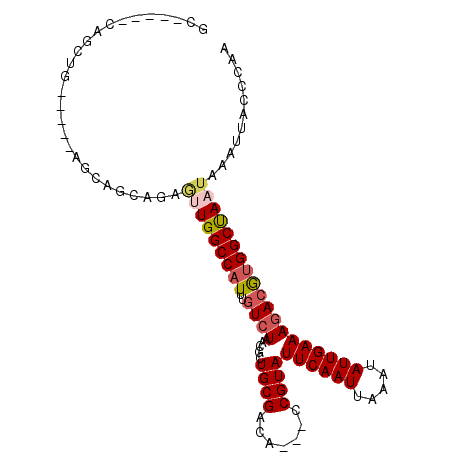
| Location | 10,683,583 – 10,683,687 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.78 |
| Mean single sequence MFE | -22.02 |
| Consensus MFE | -15.24 |
| Energy contribution | -16.18 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10683583 104 + 20766785 AGUUGGCCAUUUGUCUAACGCUGCGACA---CCGUAUUCAAUUAAAUAUUGAAAGACGUGGCUAAUAAAUUACCCAA-----ACGCA--AACGCCAAGCCGGACACAAUAGUCG .(((((((((..((((.....((((...---.))))((((((.....)))))))))))))))))))...........-----..((.--...)).......(((......))). ( -23.40) >DroPse_CAF1 23266 109 + 1 CACUGGCCAUUUGUCUGGCCCUGCGACA---CCGUAUUCAAUUAAAUAUUGAAAGACAUGGCCAAUAAAUUACCCAACACGAACACG--AACGCCAAAUGGAAUACCAAACCAG ..(((((((((((.((((((.((((...---.))))((((((.....))))))......)))))...............((....))--...).))))))).........)))) ( -23.50) >DroGri_CAF1 25762 98 + 1 CGAUGUCCAUUUAUCUA-AGCUGCGACA---GCGUAUUCAAUUAAAUAUUGAAACACGAGACUAAUAAAUUACACGA-----AAGCUCUGAGGCC---AUGAAUGCCGAU---- .............((..-((((((....---))((.((((((.....))))))))......................-----.))))..))(((.---......)))...---- ( -14.90) >DroEre_CAF1 20830 101 + 1 AGUUGGCCAUUUGUCUAACGCUGCGACA---CCGUAUUCAAUUAAAUAUUGAAAGACGUGGCUAAUAAAUUACCCAA-----AGGCA--AACGCCAAGCCGAACACAAUGG--- .(((((((((..((((.....((((...---.))))((((((.....)))))))))))))))))))...........-----.(((.--...)))................--- ( -25.10) >DroYak_CAF1 21348 104 + 1 AGUUGGCCAUUUGUCUAACGCUGCGACA---CCGUAUUCAAUUAAAUAUUGAAAGACGUGGCUAAUAAAUUACCCAA-----ACGCA--AACGCCAAGCCGAACACAAUGGUCG ....((((((((((.....((((((...---.(((.((((((.....))))))..)))(((.(((....))).))).-----.....--..)))..)))...))).))))))). ( -22.20) >DroAna_CAF1 20089 106 + 1 UCUUGGCCAUUUGUCUAACGCUGCGACACCACCGUAUUCAAUUAAAUAUUGAAAGACGUGGCUAAUAAAUUACCCA------ACGGA--AACGCAAGCCCAAACACAAAUGUCG ....(((.((((((.....((((((...((((.((.((((((.....))))))..))))))...........((..------..)).--..))).)))......))))))))). ( -23.00) >consensus AGUUGGCCAUUUGUCUAACGCUGCGACA___CCGUAUUCAAUUAAAUAUUGAAAGACGUGGCUAAUAAAUUACCCAA_____ACGCA__AACGCCAAGCCGAACACAAAAGUCG .(((((((((..((((.....((((.......))))((((((.....)))))))))))))))))))................................................ (-15.24 = -16.18 + 0.95)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:13 2006