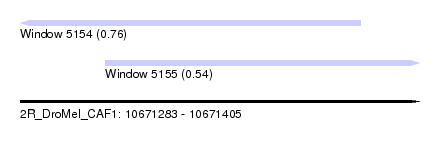
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,671,283 – 10,671,405 |
| Length | 122 |
| Max. P | 0.760370 |

| Location | 10,671,283 – 10,671,387 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 69.14 |
| Mean single sequence MFE | -15.67 |
| Consensus MFE | -5.51 |
| Energy contribution | -5.59 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
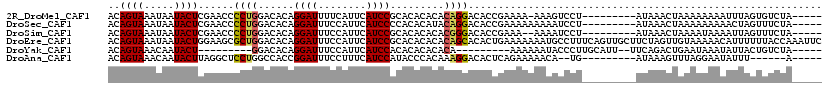
>2R_DroMel_CAF1 10671283 104 - 20766785 ACAGUAAAUAAUACUCGAACCCCUGGACACAGGAUUUUCAUUCAUCCGCACACACACAGGACACCGAAAA-AAAGUCCU---------AUAAACUAAAAAAAAUUUAGUGUCUA----- ..((((.....))))........(((((((.((((........))))..........(((((........-...)))))---------...................)))))))----- ( -17.70) >DroSec_CAF1 8165 105 - 1 ACAGUAAAUAAUACUCGAACCCCUGGACACAGGAUUUCCAUUCAUCCCCACACAUACAGGACACCGAAAAAAAAAUCCU---------AUAAACUAAAAAAAAACUAGUUUCUA----- ..((((.....)))).(((..((((....))))..)))...................((((..............))))---------..((((((.........))))))...----- ( -10.24) >DroSim_CAF1 8267 103 - 1 ACAGUAAAUAAUACUCGAACCCCUGGACACAGGAUUUCCAUUCAUCCGCACACACACGGGACACCGAAA--AAAAUCCU---------AUAAACUAAAAUAAAAUUAGUUUCUA----- ..((((.....)))).(((..((((....))))..)))..................(((....)))...--........---------..(((((((.......)))))))...----- ( -14.00) >DroEre_CAF1 8243 119 - 1 ACAGUAAAUAAUACUGGAAGCGCUGGACACAGGAUUUCCAUUCAUCCGCACACACACAGCACACUGAAAAAAAUGCCUUUCAGUUGCUUCUAGUUGUAAAAACAUUUUUUACCAAAUUC ............(((((((((((((......((((........)))).........))))..(((((((........))))))).))))))))).((((((.....))))))....... ( -28.86) >DroYak_CAF1 8479 94 - 1 ACAGUAAACAAUACU---------GGACACAGGAUUUCCAUUCAUCCACACACACACA---------AAAAAAUACCCUUGCAUU--UUCAGACUGAAUAAAUAUUACUGUCUA----- .(((((.....))))---------)((((..((((........))))...........---------..................--(((.....)))..........))))..----- ( -10.60) >DroAna_CAF1 8120 97 - 1 ACAGUAAACAAUACUUAGGCUCCUGGCCACCGGAUUUCCUUUCAUCCAUACCCACAAAGGACACUCAGAAAAACA--UG---------AUAAAGUUUAGGAAUAUUU------A----- .............(((((((((((((...))((((........))))..........))))...(((........--))---------)....))))))).......------.----- ( -12.60) >consensus ACAGUAAAUAAUACUCGAACCCCUGGACACAGGAUUUCCAUUCAUCCGCACACACACAGGACACCGAAAAAAAAAUCCU_________AUAAACUAAAAAAAAAUUAGUUUCUA_____ ..((((.....))))......((((......((((........)))).........))))........................................................... ( -5.51 = -5.59 + 0.09)

| Location | 10,671,309 – 10,671,405 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 79.85 |
| Mean single sequence MFE | -27.07 |
| Consensus MFE | -18.00 |
| Energy contribution | -19.67 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
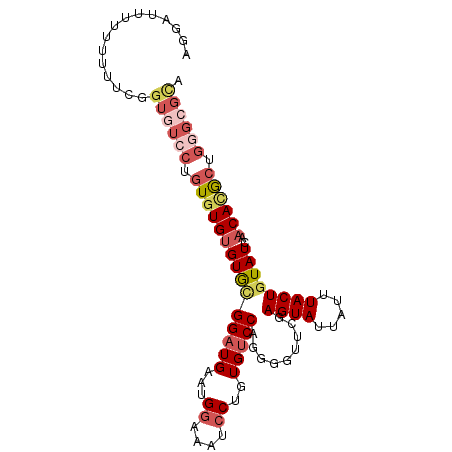
>2R_DroMel_CAF1 10671309 96 + 20766785 AGGACUUU-UUUUCGGUGUCCUGUGUGUGUGCGGAUGAAUGAAAAUCCUGUGUCCAGGGGUUCGAGUAUUAUUUACUGUAUCAACACGCUGGGAGCA ........-......((..((.((((((((((((.(((((((((..((((....))))..))......)))))))))))))..)))))).))..)). ( -25.50) >DroSec_CAF1 8191 97 + 1 AGGAUUUUUUUUUCGGUGUCCUGUAUGUGUGGGGAUGAAUGGAAAUCCUGUGUCCAGGGGUUCGAGUAUUAUUUACUGUAUCAACACGCUGGGCGCA (((((((((..((((...(((..(....)..))).)))).)))))))))((((((((.((((..((((.....)))).....))).).)))))))). ( -33.00) >DroSim_CAF1 8293 95 + 1 AGGAUUUU--UUUCGGUGUCCCGUGUGUGUGCGGAUGAAUGGAAAUCCUGUGUCCAGGGGUUCGAGUAUUAUUUACUGUAUCAACAUGCUGGGCGCA .(((....--.))).((((((.((((((((((((.(((((((((..((((....))))..)))......))))))))))))..)))))).)))))). ( -30.30) >DroEre_CAF1 8283 97 + 1 AAGGCAUUUUUUUCAGUGUGCUGUGUGUGUGCGGAUGAAUGGAAAUCCUGUGUCCAGCGCUUCCAGUAUUAUUUACUGUAUCAACACGCUGGGCGUA .(((.((((((((((.((..(.......)..))..)))).)))))))))..((((((((....(((((.....)))))........))))))))... ( -29.00) >DroYak_CAF1 8512 79 + 1 AGGGUAUUUUUU---------UGUGUGUGUGUGGAUGAAUGGAAAUCCUGUGUCC---------AGUAUUGUUUACUGUAUCAACACGCUGGGCGUA (((((.((..((---------..(.(......).)..))..)).)))))..((((---------(((..((((.........)))).)))))))... ( -17.40) >DroAna_CAF1 8140 95 + 1 CA--UGUUUUUCUGAGUGUCCUUUGUGGGUAUGGAUGAAAGGAAAUCCGGUGGCCAGGAGCCUAAGUAUUGUUUACUGUAUCAACACACUGGGCGCC ..--...........((((((..(((((((.(((((........)))))...)))..((.....((((.....))))...))..))))..)))))). ( -27.20) >consensus AGGAUUUUUUUUUCGGUGUCCUGUGUGUGUGCGGAUGAAUGGAAAUCCUGUGUCCAGGGGUUCGAGUAUUAUUUACUGUAUCAACACGCUGGGCGCA ...............((((((.(((((((((((((((...((....))..))))).........((((.....))))))))..)))))).)))))). (-18.00 = -19.67 + 1.67)

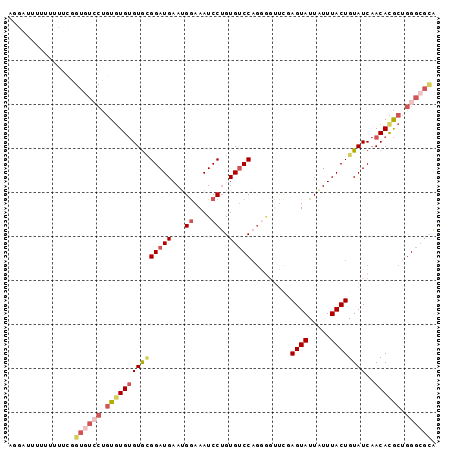
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:07 2006