| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,670,739 – 10,670,853 |
| Length | 114 |
| Max. P | 0.682308 |

| Location | 10,670,739 – 10,670,853 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.19 |
| Mean single sequence MFE | -32.54 |
| Consensus MFE | -21.73 |
| Energy contribution | -22.57 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
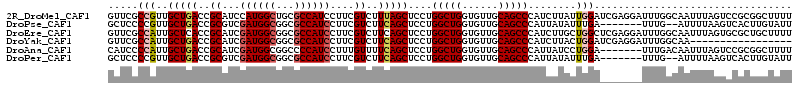
>2R_DroMel_CAF1 10670739 114 + 20766785 AAAAGCCGCGGACUAAAUUGCCAAAUCCUCGAUCCAAUAAGAUGGGCUGCAACACCAGCCAGGAGCUAAAGACGAAGGAUGGCGCAGCCAUGGAUGCGGUCAGCAACGGCGAAC ....((((..((((.....((((..((((((.((..........(((((......)))))..........)))).))))))))(((.(....).))))))).....)))).... ( -35.05) >DroPse_CAF1 9982 105 + 1 AAUACAAGUGACUUAAAAU--CAAA-------UCAAAUAUAAUGGGCUGCAACACCAGCCAGGAGCUGAAGACGAAGGAUGGCGCCGCCAUCGACGCGGUCAGCAACGGGGAGC .......((..(((.....--....-------............(((((......))))).(..(((((...((...((((((...))))))....)).)))))..))))..)) ( -28.40) >DroEre_CAF1 7583 114 + 1 AAAAGCAGCGCACUAAAUUGCCAAAUCCUCGAGCCAGCAAGAUGGGCUGCAACACCAGCCAGGAGCUGAAGACGAAGGAUGGCGCCGCCAUCGAUGCGGUGAGCAAUGGCGAAC ........(((.....(((((...(((((((...((((......(((((......)))))....))))....)).)))))..((((((.......)))))).))))).)))... ( -36.80) >DroYak_CAF1 7918 97 + 1 -----------------UUGCCAAAUCCUCGAUCCAGUAAGAUGGGCUGCAACACCAGCCAGGAGCUGAAGACGAAGGAUGGCGCCGCCAUCGAUGCGGUCAGCAAUGGCGAAC -----------------((((((..((((..(((......))).(((((......)))))))))(((((...((...((((((...))))))....)).)))))..)))))).. ( -37.20) >DroAna_CAF1 7585 107 + 1 AAAAGCCGCGGACUAAAUUGUCAAA-------UCCAGGAUAAUGGGCUGCAACACCAGCCAGGAGCUGAAAACAAAGGAUGGGGCCGCCAUCGAUGCGGUCAGCAAUGGGGAUG .....((...(((......)))...-------.(((........(((((......)))))....(((((...((...(((((.....)))))..))...)))))..)))))... ( -29.40) >DroPer_CAF1 10050 105 + 1 AAUACAAGUGACUUAAAAU--CAAA-------UCAAAUAUAAUGGGCUGCAACACCAGCCAGGAGCUGAAGACGAAGGAUGGCGCCGCCAUCGACGCGGUCAGCAACGGGGAGC .......((..(((.....--....-------............(((((......))))).(..(((((...((...((((((...))))))....)).)))))..))))..)) ( -28.40) >consensus AAAAGCAGCGAACUAAAUUGCCAAA_______UCCAAUAAAAUGGGCUGCAACACCAGCCAGGAGCUGAAGACGAAGGAUGGCGCCGCCAUCGAUGCGGUCAGCAACGGCGAAC ............................................(((((......)))))....(((((...((...((((((...))))))....)).))))).......... (-21.73 = -22.57 + 0.83)


| Location | 10,670,739 – 10,670,853 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.19 |
| Mean single sequence MFE | -34.76 |
| Consensus MFE | -20.38 |
| Energy contribution | -20.85 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10670739 114 - 20766785 GUUCGCCGUUGCUGACCGCAUCCAUGGCUGCGCCAUCCUUCGUCUUUAGCUCCUGGCUGGUGUUGCAGCCCAUCUUAUUGGAUCGAGGAUUUGGCAAUUUAGUCCGCGGCUUUU ....(((((.(((((..(((.(....).)))(((((((((.((((.........(((((......))))).........)))).)))))..))))...)))))..))))).... ( -39.67) >DroPse_CAF1 9982 105 - 1 GCUCCCCGUUGCUGACCGCGUCGAUGGCGGCGCCAUCCUUCGUCUUCAGCUCCUGGCUGGUGUUGCAGCCCAUUAUAUUUGA-------UUUG--AUUUUAAGUCACUUGUAUU .......(..(((((..(((..((((((...))))))...)))..)))))..).(((((......)))))...((((..(((-------((((--....)))))))..)))).. ( -32.20) >DroEre_CAF1 7583 114 - 1 GUUCGCCAUUGCUCACCGCAUCGAUGGCGGCGCCAUCCUUCGUCUUCAGCUCCUGGCUGGUGUUGCAGCCCAUCUUGCUGGCUCGAGGAUUUGGCAAUUUAGUGCGCUGCUUUU ...(((((((((.....)))...))))))((((((..(((((...(((((....(((((......)))))......)))))..)))))...)))).....((....)))).... ( -38.50) >DroYak_CAF1 7918 97 - 1 GUUCGCCAUUGCUGACCGCAUCGAUGGCGGCGCCAUCCUUCGUCUUCAGCUCCUGGCUGGUGUUGCAGCCCAUCUUACUGGAUCGAGGAUUUGGCAA----------------- ((.((((((((.((....)).))))))))))(((((((((.((((.........(((((......))))).........)))).)))))..))))..----------------- ( -37.67) >DroAna_CAF1 7585 107 - 1 CAUCCCCAUUGCUGACCGCAUCGAUGGCGGCCCCAUCCUUUGUUUUCAGCUCCUGGCUGGUGUUGCAGCCCAUUAUCCUGGA-------UUUGACAAUUUAGUCCGCGGCUUUU ..........(((((..(((..(((((.....)))))...)))..)))))....(((((......)))))......((((((-------((.........)))))).))..... ( -28.30) >DroPer_CAF1 10050 105 - 1 GCUCCCCGUUGCUGACCGCGUCGAUGGCGGCGCCAUCCUUCGUCUUCAGCUCCUGGCUGGUGUUGCAGCCCAUUAUAUUUGA-------UUUG--AUUUUAAGUCACUUGUAUU .......(..(((((..(((..((((((...))))))...)))..)))))..).(((((......)))))...((((..(((-------((((--....)))))))..)))).. ( -32.20) >consensus GUUCCCCAUUGCUGACCGCAUCGAUGGCGGCGCCAUCCUUCGUCUUCAGCUCCUGGCUGGUGUUGCAGCCCAUCAUACUGGA_______UUUGGCAAUUUAGUCCGCUGCUUUU .....(((..(((((..((...((((((...))))))....))..)))))....(((((......)))))........)))................................. (-20.38 = -20.85 + 0.47)
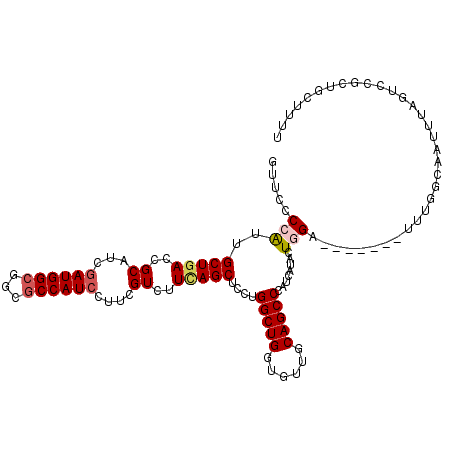
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:48:06 2006