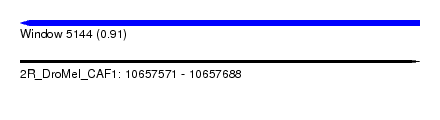
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,657,571 – 10,657,688 |
| Length | 117 |
| Max. P | 0.910251 |

| Location | 10,657,571 – 10,657,688 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.70 |
| Mean single sequence MFE | -44.22 |
| Consensus MFE | -27.25 |
| Energy contribution | -28.03 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
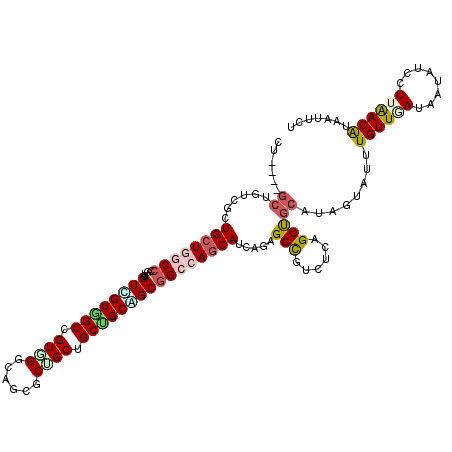
>2R_DroMel_CAF1 10657571 117 - 20766785 CUG---GCUGUUGCUGCUGACCUGUAGUCGUGGCCGUGGGCAAAGCUGCUGCUGCAGCGGCCAACAUCAGUGGCGUCUCUGCCGCAUAGUACUUGUUGAUAAUGUCCUUGACGUAGUUCU ..(---((..(.(((((......))))).)..)))((.(((...(((((....))))).))).))....((((((....))))))..((.((((((..(........)..))).))).)) ( -43.50) >DroVir_CAF1 138436 117 - 1 CUC---GCUCCCGCUGCUGACCUGUUGUUGUCGCUGUAGGCAGCGCUGCUGCGGCAGCAGCCAGCAUUAGGGGCGUCUCAGCUGCAUAGUAUUUGUUAAUAAUAUCCUUAACAUAAUUCU ..(---(((((...(((((....(((((((((((.((((......)))).))))))))))))))))...))))))..................((((((........))))))....... ( -40.60) >DroPse_CAF1 115803 117 - 1 CCA---GCUGUCUCUGCUGGCCCGUGGUGGUGGCCGUGGGCAGGGCGGCUGCUGCUGCGGCCAGCAUCAGAGGCGUCUCGGCUGCGUAGUAUUUGUUGAUGAUAUCUUUAACAUAAUUGU .((---((((((((((((((((.(..(..(..(((((.......)))))..)..)..))))))))...)))))).....))))).........((((((........))))))....... ( -51.50) >DroGri_CAF1 127441 120 - 1 CUCUCUGCUCUCUCUGCUGGCCGGUUGUCGUCGCUGUGGGCAGCGCUGCUGCAGCGGCGGCAAGCAUUAACGGUGUUUCGGCUGCAUAAUAUUUGUUUAUAAUAUCCUUAACAUAAUUCU ..............(((.((((((((((((((((((..(.((....)))..))))))))))))((((.....)))).))))))))).................................. ( -38.20) >DroMoj_CAF1 160352 109 - 1 -----------UGCUGCUGGCCCGUCGUAGUUGCUGUCGGCAGCGCUGCUGCGGCGGCGGCCAGCAUCAAAGGCGUCUCAGCUGCAUAGUAUUUGUUUAUAAUGUCUUUAACAUAAUUCU -----------.(.((((((((((((((((((((((....)))))..)))))))))..)))))))).)((((((((.......(((.......))).....))))))))........... ( -42.80) >DroAna_CAF1 125381 114 - 1 ------GCUGCUGCUGCUGGCCAGUGGUGGUGGCCGUGGGCAGUGCCGCAGCUGCCGCGGCAAGCAUUAGUGGCGUCUCCGCCGCAUAGUACUUGUUGAUGAUGUCCUUGACGUAGUUCU ------(((((.((.((((.(((.((((....))))))).)))))).)))))...(((((((((.((((((((((....)))))).)))).))))))).))(((((...)))))...... ( -48.70) >consensus CU____GCUGUCGCUGCUGGCCCGUGGUCGUGGCCGUGGGCAGCGCUGCUGCUGCAGCGGCCAGCAUCAGAGGCGUCUCAGCUGCAUAGUAUUUGUUGAUAAUAUCCUUAACAUAAUUCU ......((......((((((((....((((((((.((((......)))).)))))))))))))))).....(((......)))))........((((((........))))))....... (-27.25 = -28.03 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:57 2006