
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,656,956 – 10,657,064 |
| Length | 108 |
| Max. P | 0.864569 |

| Location | 10,656,956 – 10,657,064 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.76 |
| Mean single sequence MFE | -38.65 |
| Consensus MFE | -20.59 |
| Energy contribution | -22.48 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
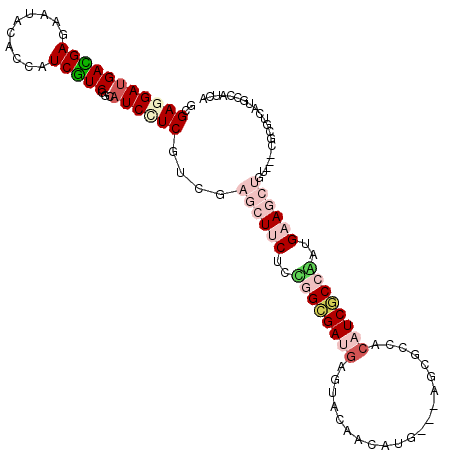
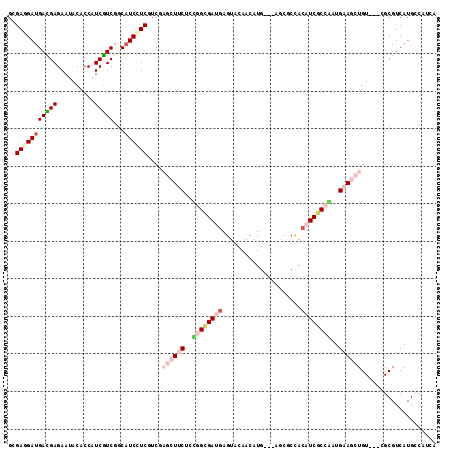
>2R_DroMel_CAF1 10656956 108 + 20766785 GUGAGGAUGACGAGAAUACACCAUCGUCUGCAUCUUCGUCGAGCUUCUCUGGCGAUGAGUUCAAUAUG---AGCGCCACAUCGCCAUUGAAGCUGU---CGCGUCAUGCCAUCA .(((((((((((.((........(((.(.........).)))(((((..((((((((.((((.....)---)))....))))))))..)))))..)---).)))))).)).))) ( -39.70) >DroVir_CAF1 137743 111 + 1 GAGAGGAUGAUGAGAAUACGCCCUCGUCGGCAUCGUCAUCCUCGUACUCGGGCGAUGAGUACAACAUGUCGGGCGCAACCUCGCCAAUGAAAUUGU---CGCGUCAUGCCAAUA ..(((((((((((......(((......))).)))))))))))(((((((.....)))))))..((((.((((((......))))...((.....)---).)).))))...... ( -40.60) >DroPse_CAF1 115185 105 + 1 GGGAGGAUGACGAGAAUACACCAUCGUCGGCAUCCUCGUCGAGCUUCUCCAGCGAUGAGUAUAACAUG---CUGUCCACAUCGCCGCUGAAGCUGU---UGCGUCAUG---UGA ..(((((((.(((((........)).))).)))))))(.((((((((..(.(((((((((((...)))---)).....)))))).)..))))))..---..)).)...---... ( -33.50) >DroMoj_CAF1 159629 111 + 1 GCGAGGAUGAUGAGAAUACACCGUCAUCGGCAUCCUCCUCCUCGUACUCGGGCGACGAGUACAACAUGUCGGGCGCUAGUUCACCCAUGAAAAUGU---CGCGUCAUGCCAACA ..(((((.((((........(((....))))))).)))))...(((((((.....)))))))..((((.((((((....((((....))))..)))---).)).))))...... ( -34.90) >DroAna_CAF1 124715 111 + 1 GCGAGGAUGACGAGAAUACACCAUCGUCCGCCUCGUCAUCGAGCUUCUCUGGUGAUGAGUUCAACAUG---AGCUCCACAUCACCACUGAAGCUGUCGUCGCGUCAUGCCGUCA ((((.(((((((((...((......))....))))))))).((((((..(((((((((((((.....)---)))))...)))))))..))))))....))))............ ( -45.80) >DroPer_CAF1 116478 105 + 1 GGGAGGAUGACGAGAAUACACCAUCGUCGGCAUCCUCGUCGAGCUUCUCCAGCGAUGAGUAUAACAUG---CUGUCCACAUCGCCGGUGAAGCUGU---UGCGUCAUG---UGA ..(((((((.(((((........)).))).)))))))(.((((((((.((.(((((((((((...)))---)).....)))))).)).))))))..---..)).)...---... ( -37.40) >consensus GCGAGGAUGACGAGAAUACACCAUCGUCGGCAUCCUCGUCGAGCUUCUCCGGCGAUGAGUACAACAUG___AGCGCCACAUCGCCAAUGAAGCUGU___CGCGUCAUGCCAUCA ..(((((((((((..........)))))...))))))....((((((..((((((((.....................))))))))..)))))).................... (-20.59 = -22.48 + 1.89)

| Location | 10,656,956 – 10,657,064 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.76 |
| Mean single sequence MFE | -40.57 |
| Consensus MFE | -21.62 |
| Energy contribution | -21.93 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
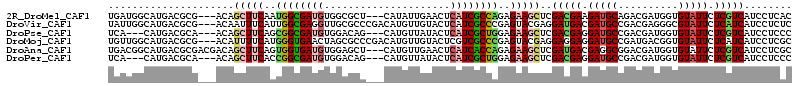
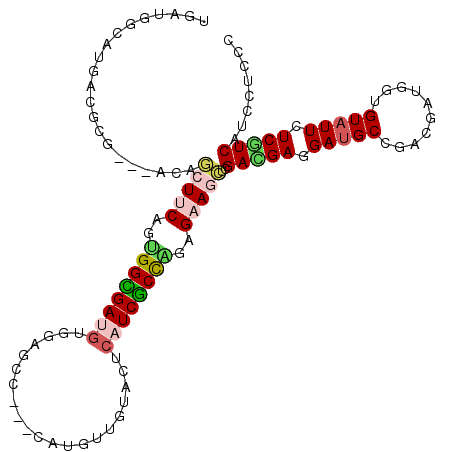
>2R_DroMel_CAF1 10656956 108 - 20766785 UGAUGGCAUGACGCG---ACAGCUUCAAUGGCGAUGUGGCGCU---CAUAUUGAACUCAUCGCCAGAGAAGCUCGACGAAGAUGCAGACGAUGGUGUAUUCUCGUCAUCCUCAC (((.((.((((((((---(..(((((..((((((((.(...(.---......)..).))))))))..)))))))).)((..(((((........)))))..)))))))))))). ( -40.00) >DroVir_CAF1 137743 111 - 1 UAUUGGCAUGACGCG---ACAAUUUCAUUGGCGAGGUUGCGCCCGACAUGUUGUACUCAUCGCCCGAGUACGAGGAUGACGAUGCCGACGAGGGCGUAUUCUCAUCAUCCUCUC ....((((((..(((---.(((((((......))))))))))....))))))((((((.......))))))((((((((.((((((......)))).....)).)))))))).. ( -40.50) >DroPse_CAF1 115185 105 - 1 UCA---CAUGACGCA---ACAGCUUCAGCGGCGAUGUGGACAG---CAUGUUAUACUCAUCGCUGGAGAAGCUCGACGAGGAUGCCGACGAUGGUGUAUUCUCGUCAUCCUCCC ...---.........---..((((((..((((((((..(((..---...))).....))))))))..)))))).((((((((((((.......).)))))))))))........ ( -37.90) >DroMoj_CAF1 159629 111 - 1 UGUUGGCAUGACGCG---ACAUUUUCAUGGGUGAACUAGCGCCCGACAUGUUGUACUCGUCGCCCGAGUACGAGGAGGAGGAUGCCGAUGACGGUGUAUUCUCAUCAUCCUCGC ....((((((....(---(.....)).((((((......)))))).))))))(((((((.....)))))))(((((.((((((((((....))))..))))))....))))).. ( -41.50) >DroAna_CAF1 124715 111 - 1 UGACGGCAUGACGCGACGACAGCUUCAGUGGUGAUGUGGAGCU---CAUGUUGAACUCAUCACCAGAGAAGCUCGAUGACGAGGCGGACGAUGGUGUAUUCUCGUCAUCCUCGC ............((((....((((((..((((((((.(.(((.---...)))...).))))))))..)))))).((((((((((...(((....)))..)))))))))).)))) ( -41.70) >DroPer_CAF1 116478 105 - 1 UCA---CAUGACGCA---ACAGCUUCACCGGCGAUGUGGACAG---CAUGUUAUACUCAUCGCUGGAGAAGCUCGACGAGGAUGCCGACGAUGGUGUAUUCUCGUCAUCCUCCC ...---.........---..((((((.(((((((((..(((..---...))).....))))))))).)))))).((((((((((((.......).)))))))))))........ ( -41.80) >consensus UGAUGGCAUGACGCG___ACAGCUUCAGUGGCGAUGUGGAGCC___CAUGUUGUACUCAUCGCCAGAGAAGCUCGACGAGGAUGCCGACGAUGGUGUAUUCUCGUCAUCCUCCC .....................(((((..((((((((.....................))))))))..)))))..(((((.(((((..........))))).)))))........ (-21.62 = -21.93 + 0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:56 2006