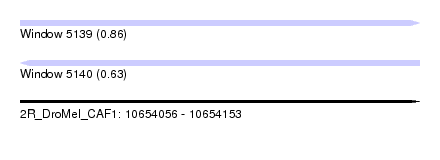
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,654,056 – 10,654,153 |
| Length | 97 |
| Max. P | 0.863595 |

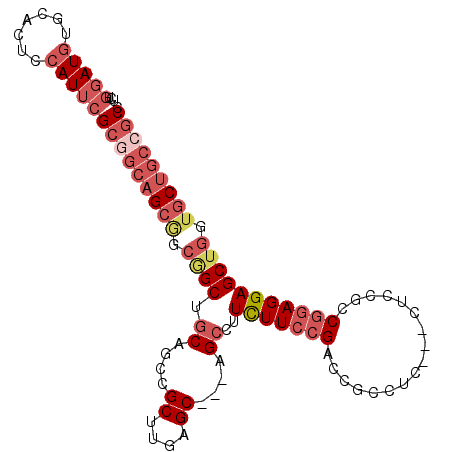
| Location | 10,654,056 – 10,654,153 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 81.63 |
| Mean single sequence MFE | -49.90 |
| Consensus MFE | -29.53 |
| Energy contribution | -32.02 |
| Covariance contribution | 2.49 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10654056 97 + 20766785 GGCGGCAGCACCAGCUCCUCCGGCGGAG---GAGGCGGUCGGAAGAAGGCC---GCCCAAGCGGCUGCAGCUGCCGCUGCGGCGAAUGGAGUGCACAUCCAGA (((((((((..((((((((((...))))---))(((((((.......))))---)))......))))..)))))))))........((((.......)))).. ( -50.40) >DroSec_CAF1 87670 97 + 1 GGCGGCAGCACCAGCUCCUCCGGCGGAG---GAGGCGGUCGGAAGAAGGCC---GCUCAAGCGGCUGCAGCCGCUGCUGCCGCGAAUGGAGUGCACAUCCAGA .((((((((.....(((((((...))))---)))((((((.......))))---))...((((((....))))))))))))))...((((.......)))).. ( -55.30) >DroSim_CAF1 88881 97 + 1 GGCGGCAGCACCAGCUCCUCCGGCGGAG---GAGGCGGUCGGAAGAAGGCC---GCUCAAGCGGCUGCAGCCGCCGCUGCCGCGAAUGGAGUGCACAUCCAGA .((((((((......((((((...))))---))(((((((....)).((((---((....))))))....)))))))))))))...((((.......)))).. ( -55.20) >DroEre_CAF1 88549 100 + 1 GGCGGCAGCACCAGCUCCUCCGGCGGAGGAGGAGGCGGUCGGAAGAAGGCU---GCUCAAGCGGCUGCAGCCGCCGCUGCCGCGAAUGGAGUGCACAUCCAGA .((((((((.....(((((((...)))))))..(((((((....)).((((---((....))))))....)))))))))))))...((((.......)))).. ( -56.20) >DroWil_CAF1 129264 82 + 1 GGCGGCA------GCUCCUCUGGCG---------GUAGUCGCAAAAAAUCUUCGGCGCAGGCUCAGGCAGCCGCUG------CUAAUGGUGUGCAUAUAGUAA ....(((------.(.((..(((((---------((.((((.((......))))))...((((.....))))))))------)))..)).))))......... ( -24.70) >DroYak_CAF1 95795 100 + 1 GGCGGCAGCACCAGCUCCUCCGGCGGCGGAGGCGGCGGUCGGAAGAAGGCU---GCUCAAGCGGCUGCGGCCGCCGCUGCCGCGAAUGGAGUGCACAUCCAAA .((((((((.......((((((....)))))).((((((((......((((---((....)))))).))))))))))))))))...((((.......)))).. ( -57.60) >consensus GGCGGCAGCACCAGCUCCUCCGGCGGAG___GAGGCGGUCGGAAGAAGGCC___GCUCAAGCGGCUGCAGCCGCCGCUGCCGCGAAUGGAGUGCACAUCCAGA .((((((((....(((..((((((.(.........).))))))....)))..........(((((....))))).))))))))...((((.......)))).. (-29.53 = -32.02 + 2.49)

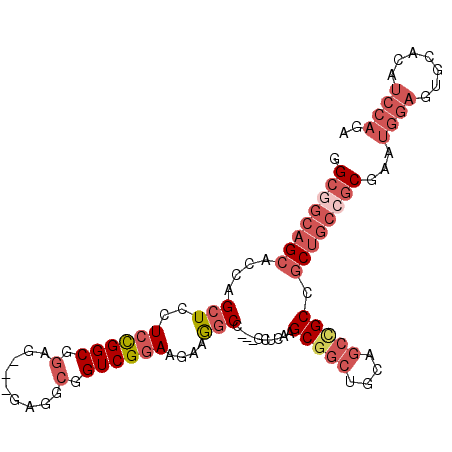
| Location | 10,654,056 – 10,654,153 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 81.63 |
| Mean single sequence MFE | -44.87 |
| Consensus MFE | -28.41 |
| Energy contribution | -30.76 |
| Covariance contribution | 2.35 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10654056 97 - 20766785 UCUGGAUGUGCACUCCAUUCGCCGCAGCGGCAGCUGCAGCCGCUUGGGC---GGCCUUCUUCCGACCGCCUC---CUCCGCCGGAGGAGCUGGUGCUGCCGCC ...(((((.......)))))......((((((((....(((((....))---))).........((((.(((---((((...))))))).)))))))))))). ( -46.50) >DroSec_CAF1 87670 97 - 1 UCUGGAUGUGCACUCCAUUCGCGGCAGCAGCGGCUGCAGCCGCUUGAGC---GGCCUUCUUCCGACCGCCUC---CUCCGCCGGAGGAGCUGGUGCUGCCGCC ...(((((.......)))))((((((((..(((..(..(((((....))---)))...)..)))((((.(((---((((...))))))).)))))))))))). ( -50.40) >DroSim_CAF1 88881 97 - 1 UCUGGAUGUGCACUCCAUUCGCGGCAGCGGCGGCUGCAGCCGCUUGAGC---GGCCUUCUUCCGACCGCCUC---CUCCGCCGGAGGAGCUGGUGCUGCCGCC ...(((((.......)))))((((((((..(((..(..(((((....))---)))...)..)))((((.(((---((((...))))))).)))))))))))). ( -50.40) >DroEre_CAF1 88549 100 - 1 UCUGGAUGUGCACUCCAUUCGCGGCAGCGGCGGCUGCAGCCGCUUGAGC---AGCCUUCUUCCGACCGCCUCCUCCUCCGCCGGAGGAGCUGGUGCUGCCGCC ...(((((.......)))))((((((((((((((....)))((....))---.)))........((((.(((((((......))))))).)))))))))))). ( -50.90) >DroWil_CAF1 129264 82 - 1 UUACUAUAUGCACACCAUUAG------CAGCGGCUGCCUGAGCCUGCGCCGAAGAUUUUUUGCGACUAC---------CGCCAGAGGAGC------UGCCGCC ....................(------(.((((((.((((.((..((((.(((....))).))).)...---------.))))).).)))------))).)). ( -22.40) >DroYak_CAF1 95795 100 - 1 UUUGGAUGUGCACUCCAUUCGCGGCAGCGGCGGCCGCAGCCGCUUGAGC---AGCCUUCUUCCGACCGCCGCCUCCGCCGCCGGAGGAGCUGGUGCUGCCGCC ...(((((.......)))))((((((((((((((....))))))..(.(---(((..(((((((...((.((....)).))))))))))))).))))))))). ( -48.60) >consensus UCUGGAUGUGCACUCCAUUCGCGGCAGCGGCGGCUGCAGCCGCUUGAGC___AGCCUUCUUCCGACCGCCUC___CUCCGCCGGAGGAGCUGGUGCUGCCGCC ...(((((.......)))))(((((((((.((((.((....((....))....))..(((((((.................))))))))))).))))))))). (-28.41 = -30.76 + 2.35)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:53 2006