| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,647,272 – 10,647,376 |
| Length | 104 |
| Max. P | 0.953480 |

| Location | 10,647,272 – 10,647,376 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 94.21 |
| Mean single sequence MFE | -24.27 |
| Consensus MFE | -21.90 |
| Energy contribution | -22.23 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
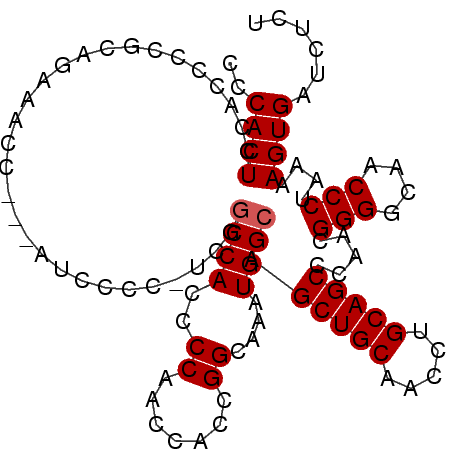
>2R_DroMel_CAF1 10647272 104 + 20766785 CCCACUCCACCCCGCACAAACCCAAAUCCCC-UCGGCCACCCCAACCACUGGCAAAUGGCAGCUGCAACCUGCAGCCCAACGGGGCAACCCUAAAAGUGAUCUCU ..((((.........................-...((((..(((.....)))....)))).(((((.....))))).....(((....)))....))))...... ( -25.40) >DroEre_CAF1 81724 101 + 1 CCCACUCCACCCCGCAGAAACC---AUCCCC-UCGACCACCCCAACCACCGGCAAAUGGCAGCUGCAACCUGCAGCCCAACGGGGCAACCCUAAAAGUGAUCUCU ......................---......-..((.(((................(((..(((((.....))))))))..(((....))).....))).))... ( -21.30) >DroYak_CAF1 88842 102 + 1 CCCACUCCACCCCGCAGCAACC---AUCCCCUUCUGCCACCCCAACCACCGGCAAAUGGCAGCUGCAACCUGCAGCCCAACGGGGCAACCCUAAAAGUGAUCUCU ..((((.......(((((..((---((.......((((............)))).))))..)))))........((((....)))).........))))...... ( -26.10) >consensus CCCACUCCACCCCGCAGAAACC___AUCCCC_UCGGCCACCCCAACCACCGGCAAAUGGCAGCUGCAACCUGCAGCCCAACGGGGCAACCCUAAAAGUGAUCUCU ..((((.............................((((..((.......))....)))).(((((.....))))).....(((....)))....))))...... (-21.90 = -22.23 + 0.33)


| Location | 10,647,272 – 10,647,376 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 94.21 |
| Mean single sequence MFE | -39.34 |
| Consensus MFE | -35.30 |
| Energy contribution | -35.08 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
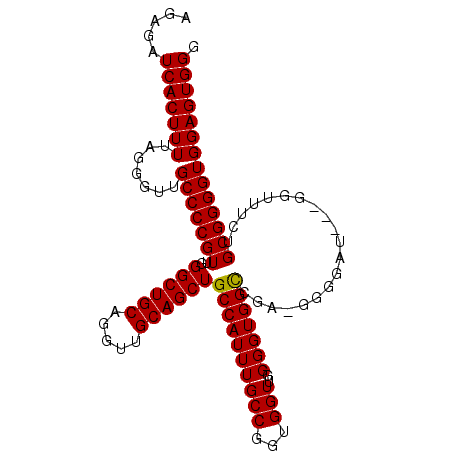
>2R_DroMel_CAF1 10647272 104 - 20766785 AGAGAUCACUUUUAGGGUUGCCCCGUUGGGCUGCAGGUUGCAGCUGCCAUUUGCCAGUGGUUGGGGUGGCCGA-GGGGAUUUGGGUUUGUGCGGGGUGGAGUGGG .....(((((((.......(((((((..((((((.....)))))).(((..(.((..(((((.....))))).-.)).)..)))......)))))))))))))). ( -40.71) >DroEre_CAF1 81724 101 - 1 AGAGAUCACUUUUAGGGUUGCCCCGUUGGGCUGCAGGUUGCAGCUGCCAUUUGCCGGUGGUUGGGGUGGUCGA-GGGGAU---GGUUUCUGCGGGGUGGAGUGGG (((((((((((((..((..((((((...((((((.....))))))(((((......)))))))))))..)).)-)))).)---)))))))............... ( -38.50) >DroYak_CAF1 88842 102 - 1 AGAGAUCACUUUUAGGGUUGCCCCGUUGGGCUGCAGGUUGCAGCUGCCAUUUGCCGGUGGUUGGGGUGGCAGAAGGGGAU---GGUUGCUGCGGGGUGGAGUGGG .....(((((((.......(((((((.(((((((.....)))))).((.(((((((.(......).))))))).))....---.....).)))))))))))))). ( -38.81) >consensus AGAGAUCACUUUUAGGGUUGCCCCGUUGGGCUGCAGGUUGCAGCUGCCAUUUGCCGGUGGUUGGGGUGGCCGA_GGGGAU___GGUUUCUGCGGGGUGGAGUGGG .....(((((((.......(((((((..((((((.....))))))((((((((((...)))..)))))))....................)))))))))))))). (-35.30 = -35.08 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:49 2006