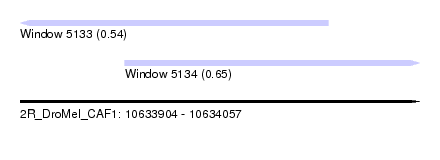
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,633,904 – 10,634,057 |
| Length | 153 |
| Max. P | 0.651158 |

| Location | 10,633,904 – 10,634,022 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.52 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -23.33 |
| Energy contribution | -23.94 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10633904 118 - 20766785 UAAACAAAUUCACCCACAUGGCAGCCUCGAUUGGGUGGCGGCAUAUCCCGCCGAUUUUCGGCGUGGGCUGAUUUUUA--GCCGCAUUCGAUUUCGAUUUCGGGUUCAAGACUUGCCAUUC .........(((((((..(((.....)))..))))))).((((..((((((((.....))))).)((((((...)))--)))(.((((((........)))))).)..))..)))).... ( -37.20) >DroPse_CAF1 101878 106 - 1 ---AUAUGUA----CAUAUG----CAUGCAUUGGGUG---GCACUGGCCGCUGUUCAGUGGCGUGGGCUGAUGUCUGAUGUCCCACCCGAAUUUGAUUUAGGGUUCAAGUCUUGCCAUUC ---..(((((----....))----)))((((((((((---(.((..((((((....))))))..((((....))))...)).))))))))((((((........))))))..)))..... ( -34.50) >DroSec_CAF1 76017 115 - 1 UAAACAAAUUCACCCACAUGGCAGCCUCGAAUGGGUG---GCAUAGCCCGCCGAUUUUCGGCGUGGGCUGAUUUUUA--GCCGCAUUCGAUUUCGAUUUCGGGUUCAAGACUUGCCGUUC .................(((((((..((((((((((.---...((((((((((.....))))..)))))).......--))).))))))).((((....))))........))))))).. ( -37.10) >DroSim_CAF1 77337 107 - 1 UAAACAAAUUCACCCACAU--------CGAAUGGGUG---GCACAGCCCGCUGAUUUUCGGCGUGGGCUGAUUUUUA--GCCGCAUUCGAUUUCGAUUUCGGGUUCAAGACUUGCCAUUC ...........(((((...--------....)))))(---(((((((((((((.....))))..)))))).......--.(((...(((....)))...)))..........)))).... ( -34.60) >DroEre_CAF1 76592 109 - 1 UAAACAGAAUCACCCACAUGGCAGCCUCGAUUGGGUG---GCACAGCCCGCCGAUUUUCGGCGUGGGCUGAUUUUUA--GCCGCAUUCGA------UUUCGGGUUCAAGACUUGCCAUUC .................((((((((((.(((((.(((---((.((((((((((.....))))..))))))(....).--)))))...)))------))..))))........)))))).. ( -36.80) >DroYak_CAF1 83752 109 - 1 UAAACAAAAGCACCCACAUGGCAGCCUCGAUUGGGUG---GCAUAGCCCGCCGAUUUUCGGCGUGGGCUGAUUUUUG--GCCGCAUUCGA------UUUCGGGUUCAAGACUUGCCAUUC .................((((((((((.(((((.(((---((.((((((((((.....))))..)))))).......--)))))...)))------))..))))........)))))).. ( -35.80) >consensus UAAACAAAUUCACCCACAUGGCAGCCUCGAUUGGGUG___GCACAGCCCGCCGAUUUUCGGCGUGGGCUGAUUUUUA__GCCGCAUUCGAUUUCGAUUUCGGGUUCAAGACUUGCCAUUC ..........((((((...............))))))...(((((((((((((.....))))..))))))............(.((((((........)))))).)......)))..... (-23.33 = -23.94 + 0.61)


| Location | 10,633,944 – 10,634,057 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 86.83 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -22.58 |
| Energy contribution | -22.92 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10633944 113 + 20766785 CUAAAAAUCAGCCCACGCCGAAAAUCGGCGGGAUAUGCCGCCACCCAAUCGAGGCUGCCAUGUGGGUGAAUUUGUUUAUAAAUACAGUGA-AACUCCGAAAGUAUGAACCGUAC ....((((..((((((((((.....)))).((....(((.............)))..))..))))))..))))(((((((.....((...-..)).(....))))))))..... ( -29.42) >DroSec_CAF1 76057 110 + 1 CUAAAAAUCAGCCCACGCCGAAAAUCGGCGGGCUAUGC---CACCCAUUCGAGGCUGCCAUGUGGGUGAAUUUGUUUAUAAAUACAGUGA-AACUCCGAGAGUAUGAACUGUCC ....((((..((((((((((.....)))).(((...((---(.(......).))).)))..))))))..))))..........(((((..-.((((...))))....))))).. ( -35.50) >DroSim_CAF1 77377 101 + 1 CUAAAAAUCAGCCCACGCCGAAAAUCAGCGGGCUGUGC---CACCCAUUCG--------AUGUGGGUGAAUUUGUUUAUAAAUACAGUGA-AACUCCGAAAGUA-GAACUGAUC ........((((((..((.(.....).))))))))...---(((((((...--------..)))))))................((((..-..((.(....).)-).))))... ( -28.30) >DroEre_CAF1 76626 110 + 1 CUAAAAAUCAGCCCACGCCGAAAAUCGGCGGGCUGUGC---CACCCAAUCGAGGCUGCCAUGUGGGUGAUUCUGUUUAUAAAUACAGUGA-AGCUGCGAGAGCACGAACUUUGC .....(((((.(((((((((.....)))).(((...((---(.(......).))).)))..))))))))))...............(..(-((.(((....)))....)))..) ( -36.50) >DroYak_CAF1 83786 111 + 1 CCAAAAAUCAGCCCACGCCGAAAAUCGGCGGGCUAUGC---CACCCAAUCGAGGCUGCCAUGUGGGUGCUUUUGUUUAUAAAUACAGUGAAAACUGCGAGAGCACAAAGUUUGC (((.....((((((.(((((.....)))))(((...))---)........).))))).....)))(((((((((..........((((....)))))))))))))......... ( -39.80) >consensus CUAAAAAUCAGCCCACGCCGAAAAUCGGCGGGCUAUGC___CACCCAAUCGAGGCUGCCAUGUGGGUGAAUUUGUUUAUAAAUACAGUGA_AACUCCGAGAGUACGAACUGUGC .........(((((..((((.....)))))))))((((...((((((..((.((...)).))))))))...((((........))))..............))))......... (-22.58 = -22.92 + 0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:47 2006