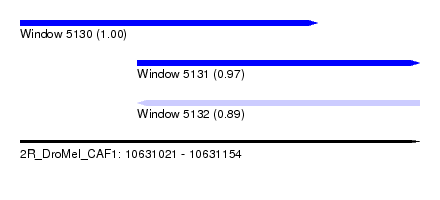
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,631,021 – 10,631,154 |
| Length | 133 |
| Max. P | 0.998589 |

| Location | 10,631,021 – 10,631,120 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 69.15 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -13.51 |
| Energy contribution | -14.23 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.51 |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.998589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
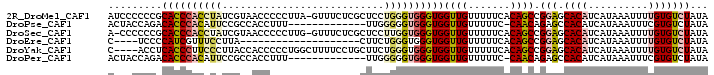
>2R_DroMel_CAF1 10631021 99 + 20766785 AUCCCCCCGCACCCACCUAUCGUAACCCCCUUA-GUUUCUCGCUCCUGGGUGGGUGGUUGUUUUUCACAGCCGGAGCACAUCAUAAAUUUUGUGUCUAUA .(((......(((((((((.((.(((.......-)))...))....)))))))))((((((.....)))))))))(((((..........)))))..... ( -27.70) >DroPse_CAF1 99218 86 + 1 ACUACCAGACACCCACAUUCCGCCACCUUU-------------UUGGGGGUGGGUGGUUGUUUUUC-CAACAGAGCCACAUCAUAAAUUUCGUGUCUAUA ......((((((.......(((((.(((..-------------..))))))))(((((((((....-.))))..)))))............))))))... ( -27.70) >DroSec_CAF1 73210 98 + 1 A-CCCCCCGCACCCACCUAUCGUAACCCCCUUG-GUUUCUCGCUCCUUGGUGGGUGGUUGUUUUUCACAGCCGGAGCACAUCAUAAAUUUUGUGUCUAUA .-....(((((((((((...((.((((.....)-)))...))......))))))))(((((.....)))))))).(((((..........)))))..... ( -28.10) >DroEre_CAF1 73727 76 + 1 C----UCCCCAUCGUUUCCUUA--------------------CUUCUGGGUGGGUGGUUGUUUUUCACAGCCGGAGCACAUCAUAAAUUUUGUGUCUAUA .----..((((((.........--------------------......)))))).((((((.....))))))(((.((((..........)))))))... ( -18.46) >DroYak_CAF1 80825 96 + 1 C----ACCUCACCCUUCCCUUACCACCCCCUGGCUUUUCCUGCUUCUGGGUGGGUGGUUGUUUUUCACAGCCGGAGCACAUCAUAAAUUUUGUGUCUAUA .----..........(((....((((((...(((.......)))...))))))..((((((.....)))))))))(((((..........)))))..... ( -28.60) >DroPer_CAF1 100493 86 + 1 ACUACCAGACACCCACAUUCCGCCACCUUU-------------UUGGGGGUGGGUGGUUGUUUUUC-CAACAGAGCCACAUCAUAAAUUUCGUGUCUAUA ......((((((.......(((((.(((..-------------..))))))))(((((((((....-.))))..)))))............))))))... ( -27.70) >consensus A___CCCCACACCCACCUAUCGCCACCCCC____________CUUCUGGGUGGGUGGUUGUUUUUCACAGCCGGAGCACAUCAUAAAUUUUGUGUCUAUA .........((((((((((...........................))))))))))((((.......)))).(((.((((..........)))))))... (-13.51 = -14.23 + 0.72)
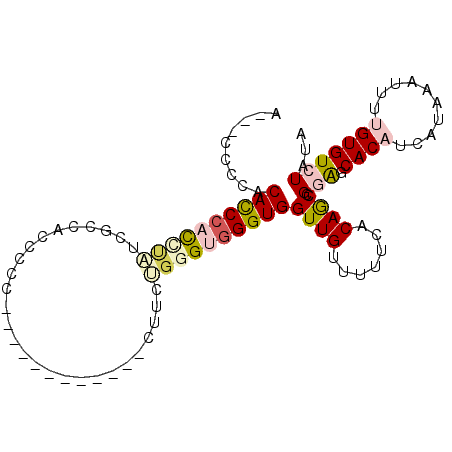
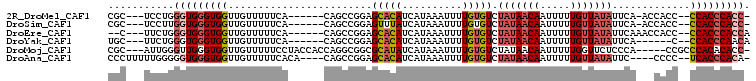
| Location | 10,631,060 – 10,631,154 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -30.59 |
| Consensus MFE | -17.57 |
| Energy contribution | -18.57 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -4.56 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10631060 94 + 20766785 CGC---UCCUGGGUGGGUGGUUGUUUUUCA------CAGCCGGAGCACAUCAUAAAUUUUGUGUCUAUAACAAUUUUUGUUAUAUUCA-ACCACC--CCACCCACC- ...---...((((((((.((((((.....)------)))))((.(((((..........))))).(((((((.....)))))))....-.))..)--)))))))..- ( -32.40) >DroSim_CAF1 74560 94 + 1 CGC---UCCUUGGUGGGUGGUUGUUUUUCA------CAGCCGGAGUUUAUCAUAAAUUUUGUGUCUAUAACAAUUUUUGUUAUAUUCA-ACCACC--CCACCCACC- ...---.....((((((((((((.......------..(((((((((((...))))))))).)).(((((((.....)))))))..))-)))).)--)))))....- ( -27.90) >DroEre_CAF1 73745 94 + 1 --C---UUCUGGGUGGGUGGUUGUUUUUCA------CAGCCGGAGCACAUCAUAAAUUUUGUGUCUAUAACAAUUUUUGUUAUAUUCAAACCACC--CCACCCACCA --.---...((((((((.((((((.....)------)))))((.(((((..........))))).(((((((.....)))))))......))..)--)))))))... ( -32.40) >DroYak_CAF1 80861 90 + 1 UGC---UUCUGGGUGGGUGGUUGUUUUUCA------CAGCCGGAGCACAUCAUAAAUUUUGUGUCUAUAACAAUUUUUGUUAUAUUCA------C--CCACCCAACA ...---...(((((((((((((((.....)------)))))((((((((..........))))).(((((((.....)))))))))))------)--)))))))... ( -33.00) >DroMoj_CAF1 137973 98 + 1 CGC---AUUGGGUUGGGUGGUUGUUUUUCCUACCACCAGGCGGCGCAUAUCAUAAAUUUUGUGUCUAUAACAAUUUUUGGUUCUCCCA-----CCGCCCACACACC- ...---....(((((((((((.(......).)))))).(((((.(.......((((..((((.......))))..)))).......).-----)))))..)).)))- ( -25.44) >DroAna_CAF1 108186 96 + 1 CCCUUUUUGGGGGUGGGUGGUUGUUUUUCACA----CAGCCGGAGCACAUCAUAAAUUUUGUGUCUAUAACAAUUUUUGUUAUAUUC----CCCC--UCACCCACA- ((((....))))((((((((((((.......)----)))))((((((((..........))))).(((((((.....))))))).))----)...--..)))))).- ( -32.40) >consensus CGC___UUCUGGGUGGGUGGUUGUUUUUCA______CAGCCGGAGCACAUCAUAAAUUUUGUGUCUAUAACAAUUUUUGUUAUAUUCA___CACC__CCACCCACC_ ...........(((((((((........................(((((..........))))).(((((((.....))))))).............))))))))). (-17.57 = -18.57 + 1.00)

| Location | 10,631,060 – 10,631,154 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 107 |
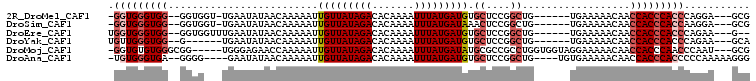
| Reading direction | reverse |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -15.26 |
| Energy contribution | -16.20 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10631060 94 - 20766785 -GGUGGGUGG--GGUGGU-UGAAUAUAACAAAAAUUGUUAUAGACACAAAAUUUAUGAUGUGCUCCGGCUG------UGAAAAACAACCACCCACCCAGGA---GCG -..(((((((--(.((((-((..(((((((.....)))))))((((((..........)))).))......------.......))))))))))))))...---... ( -32.30) >DroSim_CAF1 74560 94 - 1 -GGUGGGUGG--GGUGGU-UGAAUAUAACAAAAAUUGUUAUAGACACAAAAUUUAUGAUAAACUCCGGCUG------UGAAAAACAACCACCCACCAAGGA---GCG -(((((((((--(((.((-((....)))).....((((((((((.......)))))))))))))))((.((------(.....))).))))))))).....---... ( -29.70) >DroEre_CAF1 73745 94 - 1 UGGUGGGUGG--GGUGGUUUGAAUAUAACAAAAAUUGUUAUAGACACAAAAUUUAUGAUGUGCUCCGGCUG------UGAAAAACAACCACCCACCCAGAA---G-- .(((((((((--(.(((......(((((((.....)))))))(.((((..........))))).))).)((------(.....))).))))))))).....---.-- ( -28.70) >DroYak_CAF1 80861 90 - 1 UGUUGGGUGG--G------UGAAUAUAACAAAAAUUGUUAUAGACACAAAAUUUAUGAUGUGCUCCGGCUG------UGAAAAACAACCACCCACCCAGAA---GCA ..((((((((--(------(...(((((((.....)))))))((((((..........)))).)).((.((------(.....))).))))))))))))..---... ( -28.90) >DroMoj_CAF1 137973 98 - 1 -GGUGUGUGGGCGG-----UGGGAGAACCAAAAAUUGUUAUAGACACAAAAUUUAUGAUAUGCGCCGCCUGGUGGUAGGAAAAACAACCACCCAACCCAAU---GCG -((..((.((((((-----((((....))......(((((((((.......)))))))))..))))))))((((((.(......).))))))))..))...---... ( -32.20) >DroAna_CAF1 108186 96 - 1 -UGUGGGUGA--GGGG----GAAUAUAACAAAAAUUGUUAUAGACACAAAAUUUAUGAUGUGCUCCGGCUG----UGUGAAAAACAACCACCCACCCCCAAAAAGGG -.((((((..--((((----...(((((((.....)))))))..((((..........))))))))((.((----(.......))).)))))))).(((.....))) ( -27.60) >consensus _GGUGGGUGG__GGUG___UGAAUAUAACAAAAAUUGUUAUAGACACAAAAUUUAUGAUGUGCUCCGGCUG______UGAAAAACAACCACCCACCCAGAA___GCG .(((((((((.........................(((((((((.......))))))))).((....))..................)))))))))........... (-15.26 = -16.20 + 0.94)
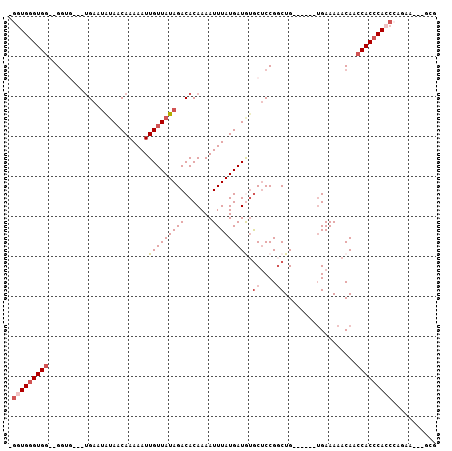
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:46 2006