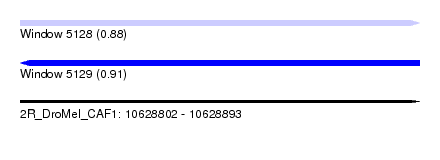
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,628,802 – 10,628,893 |
| Length | 91 |
| Max. P | 0.912918 |

| Location | 10,628,802 – 10,628,893 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 80.07 |
| Mean single sequence MFE | -22.78 |
| Consensus MFE | -14.76 |
| Energy contribution | -15.90 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
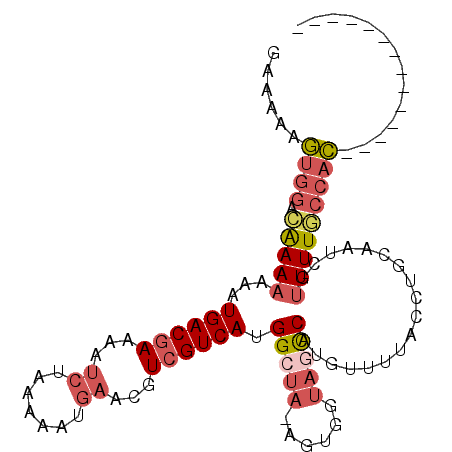
>2R_DroMel_CAF1 10628802 91 + 20766785 GAAAAAGUGGACGAAAAAAUGACGAAAAUCUAAAAAUGAACGUCGUCAUGGCUA-AGUGGUAGCCAUUUUUUACCUGCAAUCGUUUUGCCAC-------------- ......((((.(((((..(((((((...((.......))...)))))))(((((-.....)))))..................)))))))))-------------- ( -23.60) >DroPse_CAF1 97033 100 + 1 GGGAAACGAGUAGAACUACUGACCAAA---UGGAAAGGAACGUCGUCAUGGCCA-AACGGAAAUCAUGUGUCA--GGCAAUCGAUUUCACAGGUAACCACCUUAGG ((......((((....)))).(((...---(((....((......))....)))-....((((((...((...--..))...))))))...)))..))........ ( -17.10) >DroSec_CAF1 70950 91 + 1 GAAAAAGUGGACAAAAAAAUGACGAAAAUCUAAAAAUGAACGUCGUCACGGCUA-AGUGGUAGCCAUGUUUUACCUGCAAUCGUUUUGCCAC-------------- ......((((.(((((...((((((...((.......))...)))))).(((((-.....)))))..................)))))))))-------------- ( -22.90) >DroSim_CAF1 71875 91 + 1 GAAAAAGUGGAGAAAAAAAUGACGAAAAUCUAAAAAUGAACGUCGUCACGGCUA-AGUGGUAGCCAUGUUUUACCUGCAAUCGUUUUUCCAC-------------- ......(((((((((....((((((...((.......))...)))))).(((((-.....)))))..................)))))))))-------------- ( -24.60) >DroEre_CAF1 71357 91 + 1 GAAAAAGUGGACAAAAAAAUGACGAAAAUCUAAAAACGAACGUCGUCAUGGCUA-AGUGGUAGCCAUGUUUUACCUGAAAUCGUUUUGCCAC-------------- ......((((.(((((..(((((((...((.......))...)))))(((((((-.....)))))))............))..)))))))))-------------- ( -23.30) >DroYak_CAF1 78425 92 + 1 GCAAAAGUGGACAAAAAAAUGACGAAAAUCUAAAAACGAACGUCGUCAUGGGUAAAGUGGUAGCCAUGUUUUACCUGAAAUCGUUUUGCCAC-------------- ......((((.(((((..(((((((...((.......))...)))))))(((((((((((...))))..))))))).......)))))))))-------------- ( -25.20) >consensus GAAAAAGUGGACAAAAAAAUGACGAAAAUCUAAAAAUGAACGUCGUCAUGGCUA_AGUGGUAGCCAUGUUUUACCUGCAAUCGUUUUGCCAC______________ ......((((.(((((...((((((...((.......))...)))))).(((((......)))))..................))))))))).............. (-14.76 = -15.90 + 1.14)

| Location | 10,628,802 – 10,628,893 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 80.07 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -16.12 |
| Energy contribution | -16.73 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.912918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
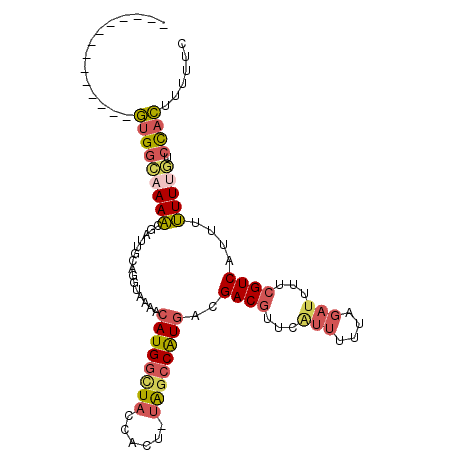
>2R_DroMel_CAF1 10628802 91 - 20766785 --------------GUGGCAAAACGAUUGCAGGUAAAAAAUGGCUACCACU-UAGCCAUGACGACGUUCAUUUUUAGAUUUUCGUCAUUUUUUCGUCCACUUUUUC --------------((((....((((.............(((((((.....-)))))))(((((...((.......))...)))))......))))))))...... ( -22.20) >DroPse_CAF1 97033 100 - 1 CCUAAGGUGGUUACCUGUGAAAUCGAUUGCC--UGACACAUGAUUUCCGUU-UGGCCAUGACGACGUUCCUUUCCA---UUUGGUCAGUAGUUCUACUCGUUUCCC ......(((((((..((.(((((((..((..--...))..)))))))))..-)))))))(((((.........((.---...))...(((....)))))))).... ( -18.00) >DroSec_CAF1 70950 91 - 1 --------------GUGGCAAAACGAUUGCAGGUAAAACAUGGCUACCACU-UAGCCGUGACGACGUUCAUUUUUAGAUUUUCGUCAUUUUUUUGUCCACUUUUUC --------------(((((((((.(((...........((((((((.....-))))))))..((((.((.......))....))))))).))))).))))...... ( -24.80) >DroSim_CAF1 71875 91 - 1 --------------GUGGAAAAACGAUUGCAGGUAAAACAUGGCUACCACU-UAGCCGUGACGACGUUCAUUUUUAGAUUUUCGUCAUUUUUUUCUCCACUUUUUC --------------(((((((((.(((...........((((((((.....-))))))))..((((.((.......))....))))))).)))).)))))...... ( -22.60) >DroEre_CAF1 71357 91 - 1 --------------GUGGCAAAACGAUUUCAGGUAAAACAUGGCUACCACU-UAGCCAUGACGACGUUCGUUUUUAGAUUUUCGUCAUUUUUUUGUCCACUUUUUC --------------(((((((((.(((...........((((((((.....-))))))))..((((.((.......))....))))))).))))).))))...... ( -25.90) >DroYak_CAF1 78425 92 - 1 --------------GUGGCAAAACGAUUUCAGGUAAAACAUGGCUACCACUUUACCCAUGACGACGUUCGUUUUUAGAUUUUCGUCAUUUUUUUGUCCACUUUUGC --------------(((((((((........((((((...(((...))).)))))).(((((((...((.......))...)))))))..))))).))))...... ( -22.50) >consensus ______________GUGGCAAAACGAUUGCAGGUAAAACAUGGCUACCACU_UAGCCAUGACGACGUUCAUUUUUAGAUUUUCGUCAUUUUUUUGUCCACUUUUUC ..............(((((((((...............((((((((......))))))))..((((...(((....)))...))))....))))).))))...... (-16.12 = -16.73 + 0.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:43 2006