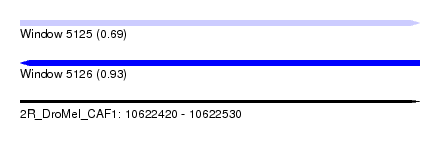
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,622,420 – 10,622,530 |
| Length | 110 |
| Max. P | 0.932481 |

| Location | 10,622,420 – 10,622,530 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.18 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -19.52 |
| Energy contribution | -20.55 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
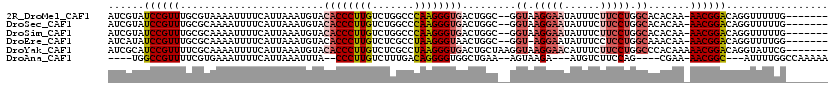
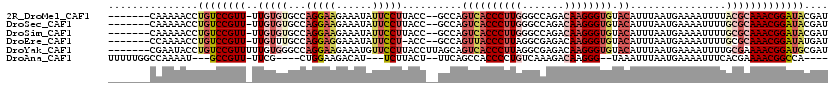
>2R_DroMel_CAF1 10622420 110 + 20766785 AUCGUAUCCGUUUGCGUAAAAUUUUCAUUAAAUGUACACCCUUGUCUGGCCCAAGGGUGACUGGC--GGUAAGGAAUAUUUCUUCCUGGCACACAA-AACGGACAGGUUUUUG------- (((...(((((((..((................((.((((((((.......))))))))))((.(--((.(((((....))))).))).)).)).)-))))))..))).....------- ( -29.70) >DroSec_CAF1 64623 110 + 1 AUCGUAUCCGUUUGCGCAAAAUUUUCAUUAAAUGUACACCCUUGUCUGGCCCAAGGGUGACUGGC--GGUAAGGAAUAUUUCUUCCUGGCACACAA-AACGGACAGGUUUUUG------- (((...(((((((.(((...((((.....))))((.((((((((.......))))))))))..))--)((.(((((......))))).)).....)-))))))..))).....------- ( -30.80) >DroSim_CAF1 65204 110 + 1 AUCGUAUCCGUUUGCGCAAAAUUUUCAUUAAAUGUACACCCUUGUCUGGCCCAAGGGUGACUGGC--GGUAAGGAAUAUUUCUUCCUGGCACACAA-AACGGACAGGUUUUUG------- (((...(((((((.(((...((((.....))))((.((((((((.......))))))))))..))--)((.(((((......))))).)).....)-))))))..))).....------- ( -30.80) >DroEre_CAF1 64911 109 + 1 AUCAUAUCCGUUUGCGCAAAAUUUUCAUUAAAUGUACACCCUUGUCUCGCCUAAGGGUAACUGGC--GGU-AGGAAUAUUUCCUCCUGGCAAACAA-AACGGACAGGUUUUGG------- ......(((((((........................(((((((.......)))))))...((.(--((.-((((.....)))).))).))....)-))))))..........------- ( -25.50) >DroYak_CAF1 71843 113 + 1 AUCGCAUCCGUUUUCGCAAAAUUUUCAUUAAAUGUACACCCUUGUCUCGCCUAAGGGUGACUGCUAAGGUAAGGAACAUUUCUUCCUGGCCCACAAAAACGGACAGGUAUUCG------- (((...((((((((.(((..((((.....))))((.((((((((.......)))))))))))))...(((.(((((......))))).)))....))))))))..))).....------- ( -30.80) >DroAna_CAF1 98650 101 + 1 ----UGGCCGUUUUCGUGAAAUUUUCAUUAAAUUUA--CCCUUGUCUUUGACAGGGGUGGCUGAA--AGUAAGA---AUGUCUUCCAG----CGAA-AACGGC---AUUUUGGCCAAAAA ----..(((((((((((((.....))))......((--((((((((...))))))))))((((.(--((.....---....))).)))----))))-))))))---.............. ( -35.90) >consensus AUCGUAUCCGUUUGCGCAAAAUUUUCAUUAAAUGUACACCCUUGUCUGGCCCAAGGGUGACUGGC__GGUAAGGAAUAUUUCUUCCUGGCACACAA_AACGGACAGGUUUUUG_______ ......((((((........................((((((((.......)))))))).........((.(((((......))))).)).......))))))................. (-19.52 = -20.55 + 1.03)
| Location | 10,622,420 – 10,622,530 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.18 |
| Mean single sequence MFE | -33.65 |
| Consensus MFE | -21.19 |
| Energy contribution | -23.53 |
| Covariance contribution | 2.34 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
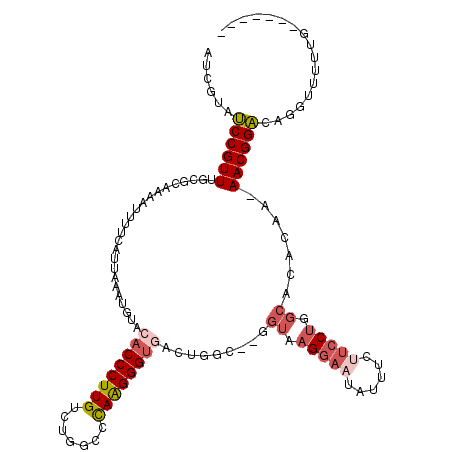
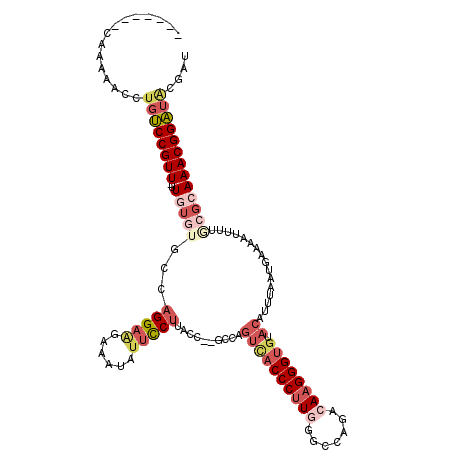
>2R_DroMel_CAF1 10622420 110 - 20766785 -------CAAAAACCUGUCCGUU-UUGUGUGCCAGGAAGAAAUAUUCCUUACC--GCCAGUCACCCUUGGGCCAGACAAGGGUGUACAUUUAAUGAAAAUUUUACGCAAACGGAUACGAU -------......(.((((((((-(.(((((..(((((......)))))....--....((((((((((.......)))))))).))...............)))))))))))))).).. ( -35.50) >DroSec_CAF1 64623 110 - 1 -------CAAAAACCUGUCCGUU-UUGUGUGCCAGGAAGAAAUAUUCCUUACC--GCCAGUCACCCUUGGGCCAGACAAGGGUGUACAUUUAAUGAAAAUUUUGCGCAAACGGAUACGAU -------......(.((((((((-(.((((((.(((((......)))))....--))..((((((((((.......)))))))).))................))))))))))))).).. ( -35.30) >DroSim_CAF1 65204 110 - 1 -------CAAAAACCUGUCCGUU-UUGUGUGCCAGGAAGAAAUAUUCCUUACC--GCCAGUCACCCUUGGGCCAGACAAGGGUGUACAUUUAAUGAAAAUUUUGCGCAAACGGAUACGAU -------......(.((((((((-(.((((((.(((((......)))))....--))..((((((((((.......)))))))).))................))))))))))))).).. ( -35.30) >DroEre_CAF1 64911 109 - 1 -------CCAAAACCUGUCCGUU-UUGUUUGCCAGGAGGAAAUAUUCCU-ACC--GCCAGUUACCCUUAGGCGAGACAAGGGUGUACAUUUAAUGAAAAUUUUGCGCAAACGGAUAUGAU -------......(.((((((((-(.........((((((.....))))-.))--(((((((((((((.(......)))))))).))....(((....))).)).))))))))))).).. ( -28.30) >DroYak_CAF1 71843 113 - 1 -------CGAAUACCUGUCCGUUUUUGUGGGCCAGGAAGAAAUGUUCCUUACCUUAGCAGUCACCCUUAGGCGAGACAAGGGUGUACAUUUAAUGAAAAUUUUGCGAAAACGGAUGCGAU -------.........(((((((((((((((..(((((......)))))..))).....(((((((((.(......)))))))).))................))))))))))))..... ( -34.50) >DroAna_CAF1 98650 101 - 1 UUUUUGGCCAAAAU---GCCGUU-UUCG----CUGGAAGACAU---UCUUACU--UUCAGCCACCCCUGUCAAAGACAAGGG--UAAAUUUAAUGAAAAUUUCACGAAAACGGCCA---- ..............---((((((-((((----(((((((....---.....))--)))))).((((.((((...)))).)))--)....................)))))))))..---- ( -33.00) >consensus _______CAAAAACCUGUCCGUU_UUGUGUGCCAGGAAGAAAUAUUCCUUACC__GCCAGUCACCCUUGGGCCAGACAAGGGUGUACAUUUAAUGAAAAUUUUGCGCAAACGGAUACGAU ...............((((((((..(((((...(((((......)))))..........((((((((((.......)))))))).))................))))))))))))).... (-21.19 = -23.53 + 2.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:40 2006