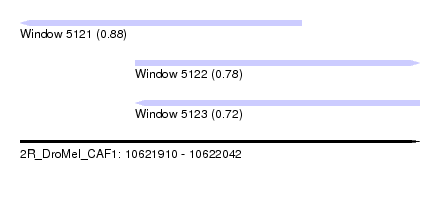
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,621,910 – 10,622,042 |
| Length | 132 |
| Max. P | 0.879721 |

| Location | 10,621,910 – 10,622,003 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 78.01 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -12.76 |
| Energy contribution | -13.09 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
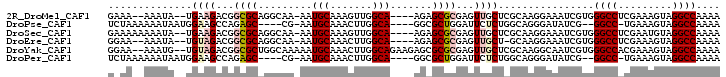
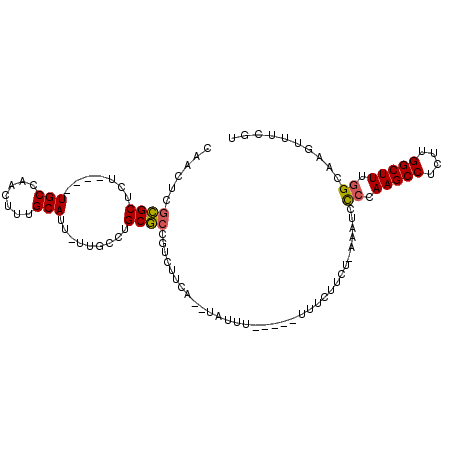
>2R_DroMel_CAF1 10621910 93 - 20766785 GAAA--AAAUA--UGAAGACGGCGCAGGCAA-AAUGCAAAGUUGGCA----AGAGCGCGAGUUGCUCGCAAGGAAAUCGUGGGCCUCGAAAGUAGGCCAAAA ....--...((--(((...((((....((..-...))...))))((.----...))(((((...))))).......)))))((((((....).))))).... ( -27.30) >DroPse_CAF1 90751 90 - 1 UCUAAAAAAUAAUGGAAGCCAGAGC----CG-AAUGCAAACUUGGCA----GGCGCUGGAUUCUCUGGCAGGGAUAUCG--GGCC-UGAAAGUAGGCCAAAA (((..............(((...((----((-(........))))).----)))((..(.....)..))..))).....--((((-((....)))))).... ( -26.40) >DroSec_CAF1 64106 95 - 1 GAAAAAAAAUA--UGAAGACGGCGCAGGCAA-AAUGCAAAGUUGGCA----AGAGCGCGAGUUGCUCGCAAGGAAAUCGUGGGCCUCGAAUGUAGGCCAAAA .........((--(((...((((....((..-...))...))))((.----...))(((((...))))).......)))))(((((.......))))).... ( -23.20) >DroEre_CAF1 64406 92 - 1 GGAA--AAAUA--UGUAGACGGCGCAGGCAA-AAUGCAAACUUGGCA----AGAGCGCGAGUUGCU-GCAAGGAAAUCGUGGGCCUCGAAAGUAGGCCAAAA ....--.....--....(.(((((((.....-..))).((((((((.----...)).)))))))))-))............((((((....).))))).... ( -27.40) >DroYak_CAF1 71323 98 - 1 GGAA--AAAUG--UGUAGACGGCGCUGGCAAAAAUGCAAACUUGGCAGAAGAGCGCGCGAGUUGCUCGCAAGGCAAUCGUGGGCCACGAAAGUAGGCCAAAA ....--.....--........(((((.(((....)))...(((.....))))))))((((.((((.(....))))))))).((((((....)).)))).... ( -33.00) >DroPer_CAF1 91948 90 - 1 UCUAAAAAAUAAUGGAAGCCAGAGC----CG-AAUGCAAACUUGGCA----GGCGCUGGAUUCUCUGGCAGGGAUAUCG--GGCC-UGAAAGUAGGCCAAAA (((..............(((...((----((-(........))))).----)))((..(.....)..))..))).....--((((-((....)))))).... ( -26.40) >consensus GAAA__AAAUA__UGAAGACGGCGCAGGCAA_AAUGCAAACUUGGCA____AGAGCGCGAGUUGCUCGCAAGGAAAUCGUGGGCCUCGAAAGUAGGCCAAAA ..............((((...((((.........(((.......))).......))))...))))................((((.........)))).... (-12.76 = -13.09 + 0.33)
| Location | 10,621,948 – 10,622,042 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 82.47 |
| Mean single sequence MFE | -21.82 |
| Consensus MFE | -14.47 |
| Energy contribution | -14.39 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
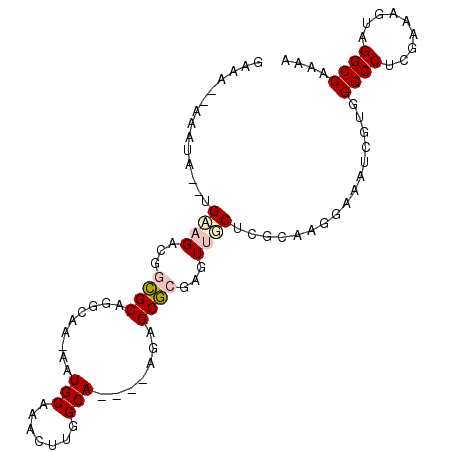
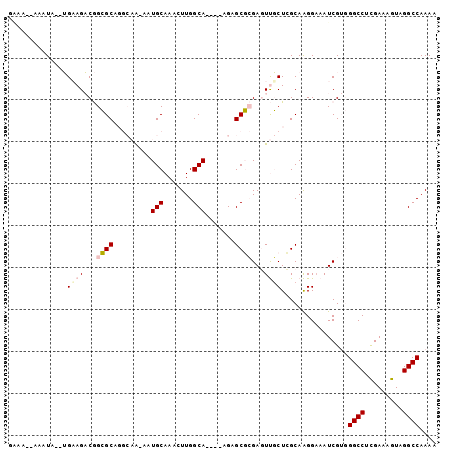
>2R_DroMel_CAF1 10621948 94 + 20766785 CAACUCGCGCUCU----UGCCAACUUUGCAUU-UUGCCUGCGCCGUCUUCA--UAUUU-----UUUCUUCC-AAAUCCCCAAGCCUCUUGGCUUUGGCAAGUUUCGU ......(((..((----(((((.....(((..-.....)))..........--.....-----........-........(((((....))))))))))))...))) ( -20.50) >DroSec_CAF1 64144 96 + 1 CAACUCGCGCUCU----UGCCAACUUUGCAUU-UUGCCUGCGCCGUCUUCA--UAUUUU---UUUUCUUCU-AAAUCCCCAAGCCUCUUGGCUUUGGCAAGUUUCGU ......(((..((----(((((.....(((..-.....)))..........--......---.........-........(((((....))))))))))))...))) ( -20.50) >DroSim_CAF1 64716 99 + 1 CAACUCGCGCUCU----UGCCAACUUUGCAUU-UUGCCUGCGCCGUCUUCA--UAUUUUUUUUUUUCUUCU-AAAUCCCCAAGCCUCUUGGCUUUGGCAAGUUUCGU ......(((..((----(((((.....(((..-.....)))..........--..................-........(((((....))))))))))))...))) ( -20.50) >DroEre_CAF1 64443 94 + 1 CAACUCGCGCUCU----UGCCAAGUUUGCAUU-UUGCCUGCGCCGUCUACA--UAUUU-----UUCCUUCU-AAAUCCCCAAGCCUCUUGGCUUUGGCAAGUUUCGC ......(((..((----((((((((..((...-..))..))..........--.....-----........-.........((((....))))))))))))...))) ( -23.10) >DroYak_CAF1 71361 99 + 1 CAACUCGCGCGCUCUUCUGCCAAGUUUGCAUUUUUGCCAGCGCCGUCUACA--CAUUU-----UUCCUUCU-AAAUCCCCAAGCCUCUUGGCUUUGGCAAGUUUCGU .((((.((((((......)).......(((....)))..))))........--.....-----........-.....((.(((((....))))).))..)))).... ( -19.90) >DroPer_CAF1 91983 95 + 1 AGAAUCCAGCGCC----UGCCAAGUUUGCAUU-CG----GCUCUGGCUUCCAUUAUUUU---UUAGAUGCUGCAAGCUGCAAGCCCCUUGGCUUUGGCAAGUUUUCC .(((..(...(((----.((((((...((...-((----(((...((...((((.....---...))))..)).)))))...))..))))))...)))..)..))). ( -26.40) >consensus CAACUCGCGCUCU____UGCCAACUUUGCAUU_UUGCCUGCGCCGUCUUCA__UAUUU_____UUUCUUCU_AAAUCCCCAAGCCUCUUGGCUUUGGCAAGUUUCGU ......((((.......(((.......))).........))))..................................((.(((((....))))).)).......... (-14.47 = -14.39 + -0.08)


| Location | 10,621,948 – 10,622,042 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 82.47 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -16.60 |
| Energy contribution | -17.16 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10621948 94 - 20766785 ACGAAACUUGCCAAAGCCAAGAGGCUUGGGGAUUU-GGAAGAAA-----AAAUA--UGAAGACGGCGCAGGCAA-AAUGCAAAGUUGGCA----AGAGCGCGAGUUG ......((((((((.(((......(((.(..((((-........-----)))).--).)))..)))(((.....-..)))....))))))----))(((....))). ( -23.40) >DroSec_CAF1 64144 96 - 1 ACGAAACUUGCCAAAGCCAAGAGGCUUGGGGAUUU-AGAAGAAAA---AAAAUA--UGAAGACGGCGCAGGCAA-AAUGCAAAGUUGGCA----AGAGCGCGAGUUG ......((((((.(((((....)))))........-.........---......--....(((...(((.....-..)))...)))))))----))(((....))). ( -23.20) >DroSim_CAF1 64716 99 - 1 ACGAAACUUGCCAAAGCCAAGAGGCUUGGGGAUUU-AGAAGAAAAAAAAAAAUA--UGAAGACGGCGCAGGCAA-AAUGCAAAGUUGGCA----AGAGCGCGAGUUG ......((((((.(((((....)))))........-..................--....(((...(((.....-..)))...)))))))----))(((....))). ( -23.20) >DroEre_CAF1 64443 94 - 1 GCGAAACUUGCCAAAGCCAAGAGGCUUGGGGAUUU-AGAAGGAA-----AAAUA--UGUAGACGGCGCAGGCAA-AAUGCAAACUUGGCA----AGAGCGCGAGUUG (((...((((((((((((....)))))........-........-----.....--((((....((....))..-..))))....)))))----))..)))...... ( -27.00) >DroYak_CAF1 71361 99 - 1 ACGAAACUUGCCAAAGCCAAGAGGCUUGGGGAUUU-AGAAGGAA-----AAAUG--UGUAGACGGCGCUGGCAAAAAUGCAAACUUGGCAGAAGAGCGCGCGAGUUG ....(((((((..(((((....)))))........-........-----.....--........(((((.(((....)))...(((.....))))))))))))))). ( -25.00) >DroPer_CAF1 91983 95 - 1 GGAAAACUUGCCAAAGCCAAGGGGCUUGCAGCUUGCAGCAUCUAA---AAAAUAAUGGAAGCCAGAGC----CG-AAUGCAAACUUGGCA----GGCGCUGGAUUCU ((((..(.((((...((((((..((...(.((((...((.((((.---.......)))).))..))))----.)-...))...)))))).----))))..)..)))) ( -30.00) >consensus ACGAAACUUGCCAAAGCCAAGAGGCUUGGGGAUUU_AGAAGAAA_____AAAUA__UGAAGACGGCGCAGGCAA_AAUGCAAACUUGGCA____AGAGCGCGAGUUG ....(((((.((.(((((....))))).))..................................((((.........(((.......))).......))))))))). (-16.60 = -17.16 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:37 2006