| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,611,945 – 10,612,050 |
| Length | 105 |
| Max. P | 0.705612 |

| Location | 10,611,945 – 10,612,050 |
|---|---|
| Length | 105 |
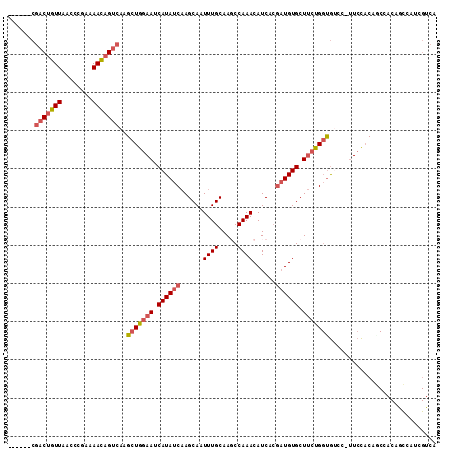
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 82.28 |
| Mean single sequence MFE | -23.17 |
| Consensus MFE | -16.42 |
| Energy contribution | -17.25 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.85 |
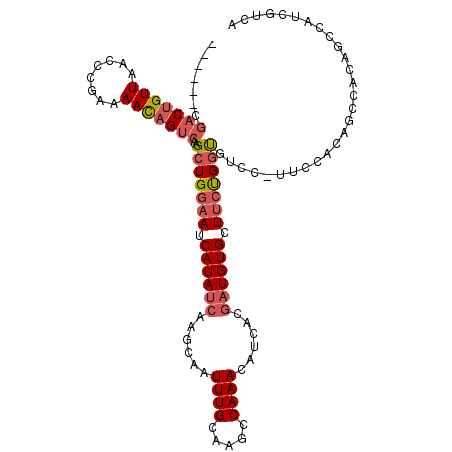
| Structure conservation index | 0.71 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10611945 105 - 20766785 ------CGACUGUUAACCCGAAAAUAGUCAAGCUGGAAUCAUAUCAAGCAAUUUGCAAGCCAAACAUCACGAUGUGCUUCUGGUGUCC-UUCCACAGCCACAGCCAUCGUCA ------.(((((((........)))))))..(((.((......)).)))...................((((((.(((..((((((..-....))).))).))))))))).. ( -25.00) >DroSec_CAF1 53972 105 - 1 ------CGACUGUUAACCCGAAAACAGUCAAGCUGGAAUCAUAUCAAGCAAUUUGCAAGCCAAACAUCACGAUGUGCUUCUGGCGUCC-UUCCACAGCCACAGCCAUCGUCA ------.(((((((........)))))))..(((.((......)).)))...................((((((.(((..((((....-.......)))).))))))))).. ( -28.50) >DroSim_CAF1 53237 105 - 1 ------CGACUGUUAACCCAAAAACAGUCAAGCUGGAAUCAUAUCAAGCAAUUUGCAAGCCAAACAUCACGAUGUGCUUCUGGUGUCC-UUCCACAGCCACAGCCAUCGUCA ------.(((((((........)))))))..(((.((......)).)))...................((((((.(((..((((((..-....))).))).))))))))).. ( -26.90) >DroEre_CAF1 52841 104 - 1 ------CGACUGUUAACCCGAAAACAGACAAGCUGGAAUCAUAUCAAGCAAUUUGCAAUCCAAACAUCACGAUGUGCUUCCGGUGACCC-UCCAC-UCCACCAUCAUCGUCA ------((((((((........)))))....(((((((.((((((......((((.....))))......)))))).))))))).....-.....-..........)))... ( -20.50) >DroYak_CAF1 61177 106 - 1 ------CGACUGUUAGCCCGAAAACAGUCAAGCUGGAAUCAUAUCCAGCAAUUUGCAAUCCAAACAUCACGAUGUGCUACUGGUGCUCCUUCCACAACCACCAUCAUGGUCA ------.(((((((........)))))))..((((((......))))))................((((.((((((....(((........)))....)).)))).)))).. ( -27.10) >DroAna_CAF1 88334 91 - 1 UGGGAACAACUGUUAACCCGAAAACUGUCAAGUUGAAAUCAUAUCAAGCAAUUUGCAAGCCAAACAUCG---UGUGCUUCCG-UAUCC-UUCUCCG---------------- .((((((....)))..)))(((....(.(((((((.............))))))))((((((.......---)).))))...-.....-)))....---------------- ( -11.02) >consensus ______CGACUGUUAACCCGAAAACAGUCAAGCUGGAAUCAUAUCAAGCAAUUUGCAAGCCAAACAUCACGAUGUGCUUCUGGUGUCC_UUCCACAGCCACAGCCAUCGUCA .......(((((((........)))))))..(((((((.((((((......((((.....))))......)))))).)))))))............................ (-16.42 = -17.25 + 0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:35 2006