| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,054,394 – 2,054,494 |
| Length | 100 |
| Max. P | 0.884452 |

| Location | 2,054,394 – 2,054,494 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.05 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -10.73 |
| Energy contribution | -11.57 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
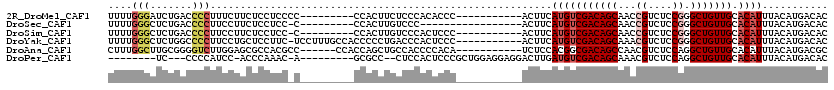
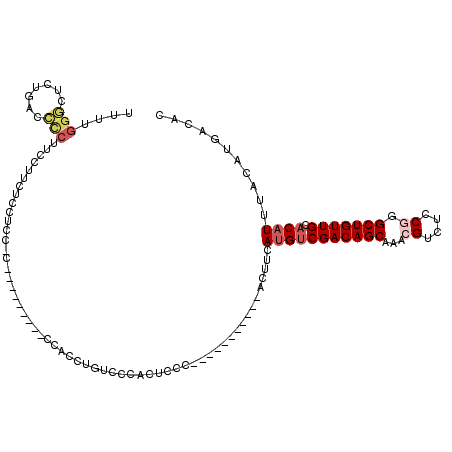
>2R_DroMel_CAF1 2054394 100 + 20766785 UUUUGGGAUCUGACCCCUUUCUUCUCCUCCCC---------CCACUUCUCCCACACCC-----------ACUUCAUGUCGACAGCAACCGUCUCCGGGCUGUUGCACAUUUACAUGACAC ....((((...((.........))...)))).---------.................-----------...(((((((((((((..(((....)))))))))).......))))))... ( -19.81) >DroSec_CAF1 39321 93 + 1 UUUUGGGCUCUGACCCCUUCCUUCUCCUCC-C---------CCACUUGUCCC-----------------ACUUCAUGUCGACAGCAACCGUCUCCGGGCUGUUGCACAUUUACAUGACAC ....(((......)))..............-.---------.....((((..-----------------.....(((((((((((..(((....)))))))))).))))......)))). ( -20.82) >DroSim_CAF1 39932 99 + 1 UUUUGGGCUCUGACCCCUUCCUUCUCCUCC-C---------CCACUUGUCCCACUCCC-----------ACUUCAUGUCGACAGCAACCGUCUCCGGGCUGUUGCACAUUUACAUGACAC ...((((....(((................-.---------......))).....)))-----------)..(((((((((((((..(((....)))))))))).......))))))... ( -19.81) >DroYak_CAF1 41864 108 + 1 UUUUGGGCUCUGGCCCCUUCCUGCUCCUUC-UCCUUUGCCACCCCCUGACCCACUCCC-----------ACUUCAUGUCGACAGCAAACGUCUCCGGGCUGUUGCACAUUUACAUGACAC ....(((...((((................-......))))..)))............-----------...(((((((((((((...((....)).))))))).......))))))... ( -21.86) >DroAna_CAF1 40703 103 + 1 CUUUGGCUUGCGGGGUCUUGGAGCGCCACGCC------CCACCAGCUGCCACCCCACA-----------UCUCCACGGCGACAGCCAACGUCUCCAGGCUGUUGCACAUUUACAUGACGC ...((((..(((((((..(((....))).)))------))....)).)))).....((-----------(.......(((((((((..........)))))))))........))).... ( -33.36) >DroPer_CAF1 50851 96 + 1 --------UC---CCCCAUCC-ACCCAAAC-A---------GCGCC--CUCCACUCCCGCUGGAGGAGGACUUGAUGUCGACAGCAAACGUCUCCAGGCUGUUGCACAUUUACAUGACAC --------..---.....(((-.((....(-(---------(((..--.........)))))..)).)))...((((((((((((............))))))).))))).......... ( -23.40) >consensus UUUUGGGCUCUGACCCCUUCCUUCUCCUCC_C_________CCACCUGUCCCACUCCC___________ACUUCAUGUCGACAGCAAACGUCUCCGGGCUGUUGCACAUUUACAUGACAC ....(((.......))).........................................................(((((((((((...((....)).))))))).))))........... (-10.73 = -11.57 + 0.84)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:08 2006