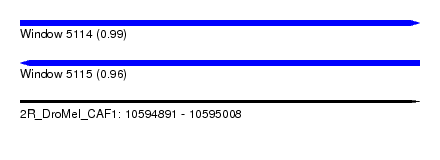
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,594,891 – 10,595,008 |
| Length | 117 |
| Max. P | 0.990556 |

| Location | 10,594,891 – 10,595,008 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.76 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -22.83 |
| Energy contribution | -23.07 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10594891 117 + 20766785 AUACGAAAGGUA---ACUGCAUGACUACGAGAAGUUAAUUAUACUGAGACACUUCCUAAUUGGCUUCCUGUUUGGGAAGCAAUCUAUUGUGCUACAUUAAUUAACUCCAAAUCGUAGUCA ...(....)...---......(((((((((..(((((((((.....((.(((..........(((((((....)))))))........)))))....))))))))).....))))))))) ( -35.47) >DroSec_CAF1 43929 117 + 1 AUACGAAAGGUA---ACUGCAUGACUACGAGAAGUUAAUUAGACUGAGACACUUCCUAAUUGGCUUCCUGUUUGGGAAGCAAUCUAUUGUGCCACAGUAAUUAACUGCAAAUCGUUGUCA ...(....)...---......((((.((((..((((((((..((((.(.(((..........(((((((....)))))))........))).).)))))))))))).....)))).)))) ( -33.37) >DroSim_CAF1 43213 117 + 1 AUAAGAAAGGUA---ACUGCAUGACUACGAGAAGAUAAUUAGACUGAGACACUUCCUAAUUGGCUUCCUGUUUGGGAAGCAAUCUAUUGUGCCACAGUAAUUAACUGCAAAUCGUUGUCA ............---......((((.((((..((.(((((..((((.(.(((..........(((((((....)))))))........))).).))))))))).)).....)))).)))) ( -27.77) >DroEre_CAF1 43193 96 + 1 AUA------------------------CGAGAAGUUAAUUAGAGUGAGACACUUCCUAAUUGGCUUCCUGUUUAGGCAGCCAUAUAUUGUGCUGUAGUGAUUAACUACAAAUCGUAGUCA .((------------------------(((((((((((((((((((...))))..)))))))))))).(((....(((((((.....)).)))))(((.....))))))..))))).... ( -27.20) >DroYak_CAF1 50617 120 + 1 AUACGAUAGGUACUCACUACAUGACUACGAGAAGUUAAUUAGACUGAGACACUUCCCAAUCGGCUUCCUGCUUAGGAAGCAAUCUAUUGUGCUGUACUGAUUAACUACAAAUCGUAGUCA ....(.(((.......)))).(((((((((..((((((((((((..((.(((..........(((((((....)))))))........))))))).)))))))))).....))))))))) ( -36.07) >consensus AUACGAAAGGUA___ACUGCAUGACUACGAGAAGUUAAUUAGACUGAGACACUUCCUAAUUGGCUUCCUGUUUGGGAAGCAAUCUAUUGUGCUACAGUAAUUAACUACAAAUCGUAGUCA ......................(((.((((..(((((((((...((.(.(((..........(((((((....)))))))........))))))...))))))))).....)))).))). (-22.83 = -23.07 + 0.24)

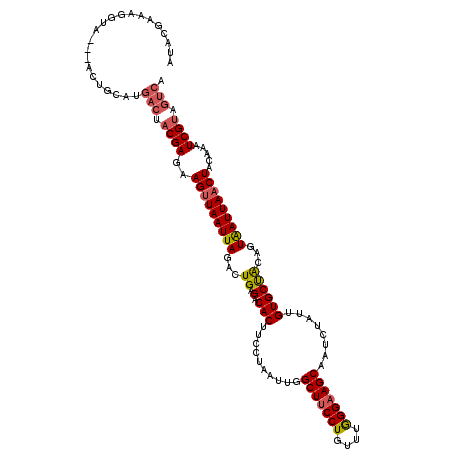
| Location | 10,594,891 – 10,595,008 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.76 |
| Mean single sequence MFE | -31.97 |
| Consensus MFE | -19.52 |
| Energy contribution | -21.36 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10594891 117 - 20766785 UGACUACGAUUUGGAGUUAAUUAAUGUAGCACAAUAGAUUGCUUCCCAAACAGGAAGCCAAUUAGGAAGUGUCUCAGUAUAAUUAACUUCUCGUAGUCAUGCAGU---UACCUUUCGUAU (((((((((...(((((((((((.((..((((..(.((((((((((......))))).))))).)...))))..))...))))))))))))))))))))......---............ ( -37.90) >DroSec_CAF1 43929 117 - 1 UGACAACGAUUUGCAGUUAAUUACUGUGGCACAAUAGAUUGCUUCCCAAACAGGAAGCCAAUUAGGAAGUGUCUCAGUCUAAUUAACUUCUCGUAGUCAUGCAGU---UACCUUUCGUAU ((((.((((...(.((((((((((((.(((((..(.((((((((((......))))).))))).)...))))).))))..)))))))).))))).))))......---............ ( -34.20) >DroSim_CAF1 43213 117 - 1 UGACAACGAUUUGCAGUUAAUUACUGUGGCACAAUAGAUUGCUUCCCAAACAGGAAGCCAAUUAGGAAGUGUCUCAGUCUAAUUAUCUUCUCGUAGUCAUGCAGU---UACCUUUCUUAU ((((.((((...(.((.(((((((((.(((((..(.((((((((((......))))).))))).)...))))).))))..))))).)).))))).))))......---............ ( -30.10) >DroEre_CAF1 43193 96 - 1 UGACUACGAUUUGUAGUUAAUCACUACAGCACAAUAUAUGGCUGCCUAAACAGGAAGCCAAUUAGGAAGUGUCUCACUCUAAUUAACUUCUCG------------------------UAU ....(((((.(((((((.....))))))).........(((((.(((....))).))))).....(((((...............))))))))------------------------)). ( -22.66) >DroYak_CAF1 50617 120 - 1 UGACUACGAUUUGUAGUUAAUCAGUACAGCACAAUAGAUUGCUUCCUAAGCAGGAAGCCGAUUGGGAAGUGUCUCAGUCUAAUUAACUUCUCGUAGUCAUGUAGUGAGUACCUAUCGUAU (((((((((.....(((((((...................(((((((....))))))).((((((((....))))))))..)))))))..)))))))))..................... ( -35.00) >consensus UGACUACGAUUUGCAGUUAAUUACUGUAGCACAAUAGAUUGCUUCCCAAACAGGAAGCCAAUUAGGAAGUGUCUCAGUCUAAUUAACUUCUCGUAGUCAUGCAGU___UACCUUUCGUAU ((((.((((...(.(((((((((....(((((....((((((((((......))))).))))).....))).)).....))))))))).))))).))))..................... (-19.52 = -21.36 + 1.84)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:30 2006