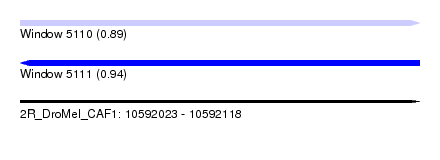
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,592,023 – 10,592,118 |
| Length | 95 |
| Max. P | 0.939555 |

| Location | 10,592,023 – 10,592,118 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 83.67 |
| Mean single sequence MFE | -21.80 |
| Consensus MFE | -15.90 |
| Energy contribution | -16.74 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10592023 95 + 20766785 CGAAAGCCACUCGUUGACACCAGAACAACUAUGUGGGCCAACAAAUGGCCCAAAAGCUGGAGCGUACUUUACCAAAAAGGACAAAAGCCAGCUCU ....(((.....((((.........))))....(((((((.....)))))))...)))(((((...((((.((.....))...))))...))))) ( -25.00) >DroSec_CAF1 41066 95 + 1 CGAAAGCCUCUCGUUGACACCAGAACAACUAUGUGGGCCAACAAAUGGCCCAAAAGCUGGAGCGCACUUUACCAAAAAGGACAAAAGCCAGCUCU ....(((.....((((.........))))....(((((((.....)))))))...)))(((((...((((.((.....))...))))...))))) ( -25.00) >DroSim_CAF1 40333 95 + 1 UGAAAGCCUCUCGUUGACACCAGAACAACUAUGUGGGCCAACAAAUGGCCCAAAAGCUGGAGCGCACUUUACCAAAAAGGACAAAAGCCAGCUCU ....(((.....((((.........))))....(((((((.....)))))))...)))(((((...((((.((.....))...))))...))))) ( -25.00) >DroEre_CAF1 40370 94 + 1 CGAAAGCCUCUCGUCGACACCACAACCACUUUGUGGGCCAGCAAAUGGCCCAAACGCCAGAGCACACUUUACCAAAAA-GACAAAAGCCAGCUCU ....(((((((.(.((.....((((.....))))((((((.....))))))...))).))))....((((.....)))-)..........))).. ( -20.30) >DroYak_CAF1 47750 74 + 1 CAA--------------------AACCACUAUGUGGGCCAACAAAAGGGCCAAAAGCUGUAGCACACUUUGCCAAAAA-GACAAAAGCCAGCUCU ...--------------------...(((...)))((((........))))...(((((..((...((((.....)))-)......))))))).. ( -13.70) >consensus CGAAAGCCUCUCGUUGACACCAGAACAACUAUGUGGGCCAACAAAUGGCCCAAAAGCUGGAGCGCACUUUACCAAAAAGGACAAAAGCCAGCUCU .................................(((((((.....)))))))......(((((...((((.((.....))...))))...))))) (-15.90 = -16.74 + 0.84)

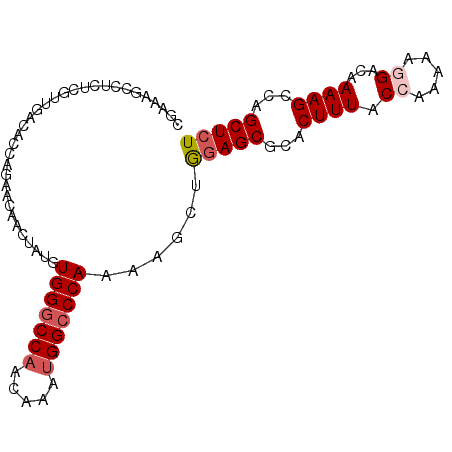
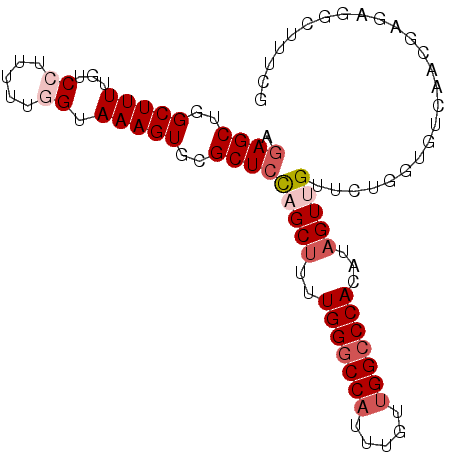
| Location | 10,592,023 – 10,592,118 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 83.67 |
| Mean single sequence MFE | -31.38 |
| Consensus MFE | -20.94 |
| Energy contribution | -22.38 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10592023 95 - 20766785 AGAGCUGGCUUUUGUCCUUUUUGGUAAAGUACGCUCCAGCUUUUGGGCCAUUUGUUGGCCCACAUAGUUGUUCUGGUGUCAACGAGUGGCUUUCG ((((((((....(((.((((.....)))).)))..)))))))).((((((((((((((((((((....)))...)).)))))))))))))))... ( -36.40) >DroSec_CAF1 41066 95 - 1 AGAGCUGGCUUUUGUCCUUUUUGGUAAAGUGCGCUCCAGCUUUUGGGCCAUUUGUUGGCCCACAUAGUUGUUCUGGUGUCAACGAGAGGCUUUCG .((((..(((((...((.....)).)))))..))))(((((..(((((((.....)))))))...)))))((((.((....)).))))....... ( -33.80) >DroSim_CAF1 40333 95 - 1 AGAGCUGGCUUUUGUCCUUUUUGGUAAAGUGCGCUCCAGCUUUUGGGCCAUUUGUUGGCCCACAUAGUUGUUCUGGUGUCAACGAGAGGCUUUCA .((((..(((((...((.....)).)))))..))))(((((..(((((((.....)))))))...)))))((((.((....)).))))....... ( -33.80) >DroEre_CAF1 40370 94 - 1 AGAGCUGGCUUUUGUC-UUUUUGGUAAAGUGUGCUCUGGCGUUUGGGCCAUUUGCUGGCCCACAAAGUGGUUGUGGUGUCGACGAGAGGCUUUCG .(((..(.((((((((-.....((((((.((.((((........))))))))))))(((((((((.....)))))).))))))))))).).))). ( -33.70) >DroYak_CAF1 47750 74 - 1 AGAGCUGGCUUUUGUC-UUUUUGGCAAAGUGUGCUACAGCUUUUGGCCCUUUUGUUGGCCCACAUAGUGGUU--------------------UUG ((((((((((((((((-.....)))))))...))..))))))).((((........))))(((...)))...--------------------... ( -19.20) >consensus AGAGCUGGCUUUUGUCCUUUUUGGUAAAGUGCGCUCCAGCUUUUGGGCCAUUUGUUGGCCCACAUAGUUGUUCUGGUGUCAACGAGAGGCUUUCG .((((..(((((...((.....)).)))))..))))(((((..(((((((.....)))))))...)))))......................... (-20.94 = -22.38 + 1.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:26 2006