
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,584,550 – 10,584,654 |
| Length | 104 |
| Max. P | 0.934505 |

| Location | 10,584,550 – 10,584,654 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 78.14 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -22.18 |
| Energy contribution | -22.18 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10584550 104 + 20766785 AUGGCCAAAUGGGUUGG-UGGGGGAAAGUGAAGAAGUUGGCGUAAGUAAGGCAGAGCCAUUUAUCAGCGUGACACCUUGCCGGGUGGCCAACUUGGAAUACUGGG .((((((......((((-..(((....((.(....((((((....))..(((...)))......)))).).)).)))..)))).)))))).((..(....)..)) ( -31.10) >DroVir_CAF1 67070 90 + 1 -AGGUAGCCGG----AC--------CGAUAGAGAAGUUGGCGUAAGUAAGGCAGCGCCAUUUAUCAGCGUGACACCUUGCCGGGUGGCCAACUUGGU--CCUGGA -......((((----((--------((.......(((((((....((((((..((((.........))))....))))))......)))))))))))--)).)). ( -30.81) >DroGri_CAF1 60237 90 + 1 -AGGUAACCAG----AC--------CGAUAGAGAAGUUGGCGUAAGUAAGGCAACGCCAUUUAUCAGCGUGACACCUUGCAGGGUGGCCAACUUGGU--UCUGGA -......((((----((--------((.......(((((((((..(((((((...)))..))))..))....(((((....))))))))))))))).--))))). ( -30.11) >DroEre_CAF1 33013 102 + 1 AUGGCCAAAUUGGA-GG--GGGGGAAAGUGAAGAAGUUGGCGUAAGUAAGGCACAGCCAUUUAUCAGCGUGACACCUUGCCGGGUGGCCAACUUGGAGUAUUGGG .((((((.......-((--.((((...((.(....((((((....))..(((...)))......)))).).)).)))).))...))))))............... ( -26.70) >DroYak_CAF1 40060 103 + 1 AUGGCCAAAUUGGUUUG--GGGGGAAAGUGAAGAAGUUGGCGUAAGUAAGGCACAGCCAUUUAUCAGCGUGACACCUUGCCGGGUGGCCAACUUGGAAUACUGGG .((((((...((((...--((((....((.(....((((((....))..(((...)))......)))).).)).))))))))..)))))).((..(....)..)) ( -27.90) >DroAna_CAF1 66894 99 + 1 AGGGGCAAAUUGGG-GGUGGUGGCCAAGUGAAGAAGUUGGCGUAAGUAAGGCAGAGCCAUUUAUCAGCGUGACACCUUGCCGGGUGGCCAACUUGGC--CCU--- ((((.(((.((((.-(.((((((((((.(.....).)))))........(((...)))...))))).)....(((((....))))).)))).))).)--)))--- ( -35.90) >consensus AUGGCCAAAUGGG__GG__GGGGGAAAGUGAAGAAGUUGGCGUAAGUAAGGCAGAGCCAUUUAUCAGCGUGACACCUUGCCGGGUGGCCAACUUGGA__ACUGGG .................................((((((((((..(((((((...)))..))))..))....(((((....)))))))))))))........... (-22.18 = -22.18 + 0.00)

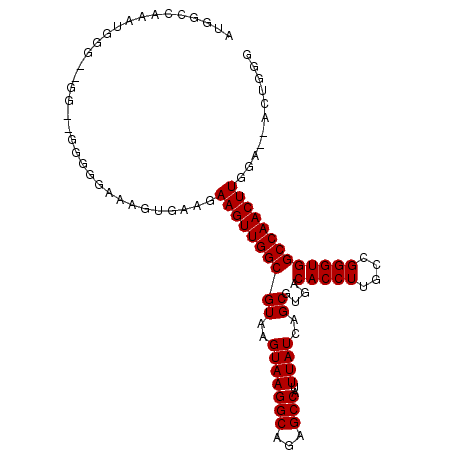

| Location | 10,584,550 – 10,584,654 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 78.14 |
| Mean single sequence MFE | -25.26 |
| Consensus MFE | -17.60 |
| Energy contribution | -17.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10584550 104 - 20766785 CCCAGUAUUCCAAGUUGGCCACCCGGCAAGGUGUCACGCUGAUAAAUGGCUCUGCCUUACUUACGCCAACUUCUUCACUUUCCCCCA-CCAACCCAUUUGGCCAU ...............((((((...((...((((......(((..((((((..............))))..))..)))........))-))...))...)))))). ( -19.88) >DroVir_CAF1 67070 90 - 1 UCCAGG--ACCAAGUUGGCCACCCGGCAAGGUGUCACGCUGAUAAAUGGCGCUGCCUUACUUACGCCAACUUCUCUAUCG--------GU----CCGGCUACCU- .((.((--(((((((((((.....((((..((((((..........))))))))))........)))))))).......)--------))----))))......- ( -28.23) >DroGri_CAF1 60237 90 - 1 UCCAGA--ACCAAGUUGGCCACCCUGCAAGGUGUCACGCUGAUAAAUGGCGUUGCCUUACUUACGCCAACUUCUCUAUCG--------GU----CUGGUUACCU- .(((((--.((((((((((......(.((((((..(((((.......))))))))))).)....)))))))).......)--------))----))))......- ( -27.81) >DroEre_CAF1 33013 102 - 1 CCCAAUACUCCAAGUUGGCCACCCGGCAAGGUGUCACGCUGAUAAAUGGCUGUGCCUUACUUACGCCAACUUCUUCACUUUCCCCC--CC-UCCAAUUUGGCCAU .........(((((((((......((.((((((((.....))....((((.(((.......))))))).......))))))))...--..-.))))))))).... ( -22.70) >DroYak_CAF1 40060 103 - 1 CCCAGUAUUCCAAGUUGGCCACCCGGCAAGGUGUCACGCUGAUAAAUGGCUGUGCCUUACUUACGCCAACUUCUUCACUUUCCCCC--CAAACCAAUUUGGCCAU .........(((((((((......((.((((((((.....))....((((.(((.......))))))).......)))))).))..--....))))))))).... ( -23.00) >DroAna_CAF1 66894 99 - 1 ---AGG--GCCAAGUUGGCCACCCGGCAAGGUGUCACGCUGAUAAAUGGCUCUGCCUUACUUACGCCAACUUCUUCACUUGGCCACCACC-CCCAAUUUGCCCCU ---(((--(.((((((((((....)))..(((((((.(.(((..((((((..............))))..))..)))).))).))))...-..))))))).)))) ( -29.94) >consensus CCCAGU__UCCAAGUUGGCCACCCGGCAAGGUGUCACGCUGAUAAAUGGCUCUGCCUUACUUACGCCAACUUCUUCACUUUCCCCC__CC__CCAAUUUGGCCAU ...........((((((((((((......))))..............(((...)))........))))))))................................. (-17.60 = -17.60 + -0.00)

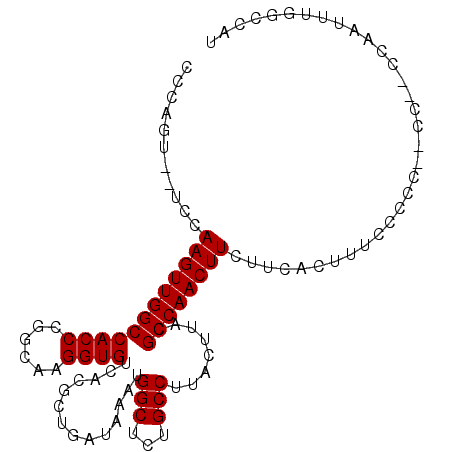
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:21 2006