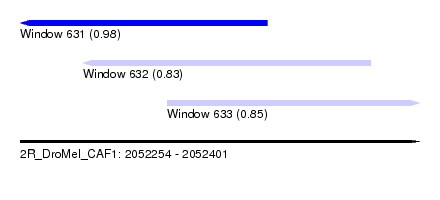
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,052,254 – 2,052,401 |
| Length | 147 |
| Max. P | 0.982605 |

| Location | 2,052,254 – 2,052,345 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 70.91 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -20.46 |
| Energy contribution | -19.02 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2052254 91 - 20766785 AUUUAUAAAUUUCUUGGCCGAGUGCCAGCGAUGUAAUCUCAUUGCUUGUAGCUUUGGCCAGC---CACUCC---------UCCACUGC-C-----UCC---GCUGCCUCCU----C .............((((((((((((.(((((((......))))))).)))..))))))))).---......---------........-.-----...---..........----. ( -22.20) >DroVir_CAF1 18199 107 - 1 AUUUAUAAAUU--CUGGCCAAAUGCCAGAGAUAUAAUCUUAUUGCUUGUAGUUUUGGUUACAUAGGACUCCAA---GGCACCAAAUAACCGAGAGUCCAAUACAGUCAGCC----C ...........--(((((.....(((((((.(((((.(.....).))))).)))))))......((((((...---((..........))..))))))......)))))..----. ( -24.50) >DroGri_CAF1 114870 92 - 1 AUUUAUAAAAU--UUGGCCAAAUGCCAGCGAUAUAAUCUUAUUGCUUGUAGUUUUGGUCACAGC-CA--CC-------ACCCAAAUAA------------UACUGUCAGCCCAAAC ...........--.(((((((((((.(((((((......))))))).)))..))))))))....-..--..-------..........------------................ ( -18.40) >DroSec_CAF1 37155 99 - 1 AUUUAUAAAUUUCUUGGCCGAGUGCCAGCGAUGUAAUCUCAUUGCUUGUAGCUUUGGCCAGC---CACUCCUCCACUCC-UCCACUGC-C-----UCC---GCUGCCUCCU----C .............((((((((((((.(((((((......))))))).)))..))))))))).---..............-........-.-----...---..........----. ( -22.20) >DroSim_CAF1 37785 99 - 1 AUUUAUAAAUUUCUUGGCCGAGUGCCAGCGAUGUAAUCUCAUUGCUUGUAGCUUUGGCCAGC---CACUCCUCCACUCC-UCCACUGC-C-----UCC---GCUGCCUCCU----C .............((((((((((((.(((((((......))))))).)))..))))))))).---..............-........-.-----...---..........----. ( -22.20) >DroEre_CAF1 36818 82 - 1 AUUUAUAAAUUUCUUGGCCGAGUGCCAGCGAUGUAAUCUCAUUGCUUGUAGCUCUGGCCAGC---CACUCCUA------------------------C---CCUUCCUCCU----C .............((((((((((((.(((((((......))))))).))).))).)))))).---........------------------------.---..........----. ( -24.20) >consensus AUUUAUAAAUUUCUUGGCCGAGUGCCAGCGAUGUAAUCUCAUUGCUUGUAGCUUUGGCCAGC___CACUCCU______C_UCCACUGC_C_____UCC___GCUGCCUCCU____C ..............(((((((((((.(((((((......))))))).))).))).)))))........................................................ (-20.46 = -19.02 + -1.44)

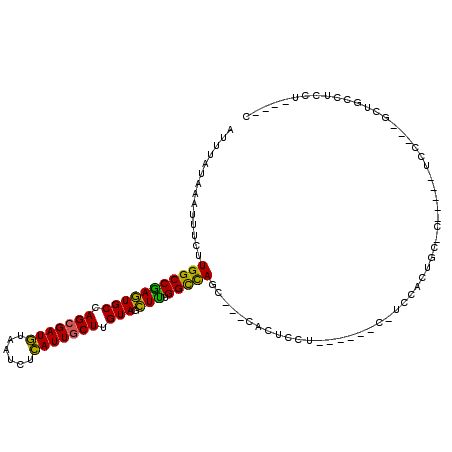
| Location | 2,052,277 – 2,052,383 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.35 |
| Mean single sequence MFE | -25.35 |
| Consensus MFE | -14.20 |
| Energy contribution | -15.46 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
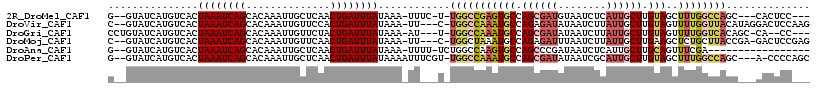
>2R_DroMel_CAF1 2052277 106 - 20766785 G--GUAUCAUGUCACUAAAUCAGCACAAAUUGCUCAACUGAUUUAUAAA-UUUC-U-UGGCCGAGUGCCAGCGAUGUAAUCUCAUUGCUUGUAGCUUUGGCCAGC---CACUCC--- (--((..........((((((((..............))))))))....-....-.-(((((((((((.(((((((......))))))).)))..))))))))))---).....--- ( -30.64) >DroVir_CAF1 18234 110 - 1 C--GUAUCAUGUCACUAAAUCAGCACAAAUUGUUCCACUGAUUUAUAAA-UU---C-UGGCCAAAUGCCAGAGAUAUAAUCUUAUUGCUUGUAGUUUUGGUUACAUAGGACUCCAAG .--(((((.......((((((((.((.....))....))))))))....-.(---(-((((.....)))))))))))..........((((.(((((((......))))))).)))) ( -25.80) >DroGri_CAF1 114896 106 - 1 CCUGUAUCAUGUCACUAAAUCAGCACAAAUUGUUCUACUGAUUUAUAAA-AU---U-UGGCCAAAUGCCAGCGAUAUAAUCUUAUUGCUUGUAGUUUUGGUCACAGC-CA--CC--- .((((..........((((((((.((.....))....))))))))....-..---.-.((((((((((.(((((((......))))))).)))..))))))))))).-..--..--- ( -26.20) >DroMoj_CAF1 63009 109 - 1 C--GUAUCAUGUCACUAAAUCAGCACAAAUUGUUCAACUGAUUUAUAAA-UU---C-UGGCUAAAUGCCAGAGAUUUAAUCUUAUUGCUUGAAGCUCUGCUUACCGA-GACUCCGAG .--............((((((((.((.....))....))))))))((((-((---(-((((.....)))))).))))).........((((.(((((........))-).)).)))) ( -22.90) >DroAna_CAF1 38759 96 - 1 G--GUAUCAUGUCACUAAAUCAGCACAAAUUGCUCAACUGAUUUAUAAA-UUUU-UCUGGCCAAGUGCCAGCCCGAUAAUCUCAUUGCUUGCAGUUUCGA----------------- .--......((((..((((((((..............))))))))....-....-.(((((.....)))))...)))).....((((....)))).....----------------- ( -15.44) >DroPer_CAF1 47873 110 - 1 G--GUAUCAUGUCACUAAAUCAGCACAAAUUGCUCAACUGAUUUAUAAAAUUUCGU-UGGCCAAAUGCCAGCGAUAUAAUCGCAUUGCUUGUAGCUUUGGCCAGC---A-CCCCAGC (--((..........((((((((..............)))))))).........((-(((((((((((.((((((........)))))).)))..))))))))))---)-))..... ( -31.14) >consensus C__GUAUCAUGUCACUAAAUCAGCACAAAUUGCUCAACUGAUUUAUAAA_UU___U_UGGCCAAAUGCCAGCGAUAUAAUCUCAUUGCUUGUAGCUUUGGUCACC___CACUCC___ ...............((((((((..............))))))))............(((((((((((.((((((........)))))).)))..)))))))).............. (-14.20 = -15.46 + 1.25)
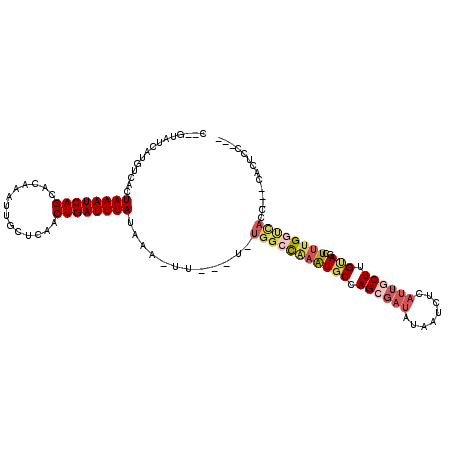
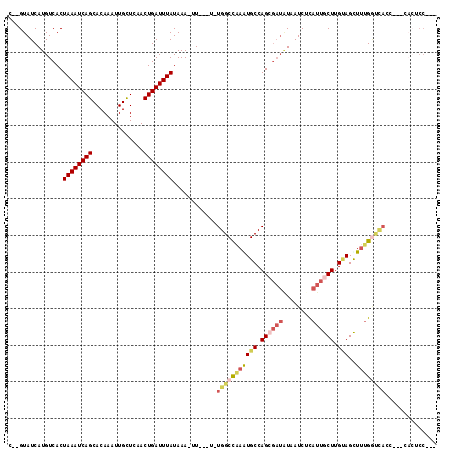
| Location | 2,052,308 – 2,052,401 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.71 |
| Mean single sequence MFE | -23.80 |
| Consensus MFE | -13.50 |
| Energy contribution | -13.92 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2052308 93 + 20766785 AUUACAUCGCUGGCACUCGGCCA-AGAAAUUUAUAAAUCAGUUGAGCAAUUUGUGCUGAUUUAGUGACAUGAUAC--CA--CAGUCGAAGCCA----GUGGA------------- ......((((((((..(((((..-.........(((((((((.((....))...)))))))))(((.........--))--).))))).))))----)))).------------- ( -29.90) >DroVir_CAF1 18271 100 + 1 AUUAUAUCUCUGGCAUUUGGCCA-G--AAUUUAUAAAUCAGUGGAACAAUUUGUGCUGAUUUAGUGACAUGAUAC--GA--A--U-GGAGCCAACUAAAGGA----CAACAGG-A .(..((((((((((.....))))-)--).....((((((((((.((....)).)))))))))).......)))).--.)--.--(-(..(((.......)).----)..))..-. ( -22.80) >DroGri_CAF1 114927 106 + 1 AUUAUAUCGCUGGCAUUUGGCCA-A--AUUUUAUAAAUCAGUAGAACAAUUUGUGCUGAUUUAGUGACAUGAUACAGGA--A--U-GGAGCCAACUAAGGGAACAACAACAGG-A (((((.((((((((.....))))-.--......((((((((((.((....)).)))))))))))))).)))))......--.--(-(...((.......))..))........-. ( -23.30) >DroYak_CAF1 39823 90 + 1 -----GUCGCUGGCACUCGGCCA-AGAAAUUUAUAAAUCAGUUGAGCAAUUUGUGCUGAUUUAGUGACAUGAUAC--CACACAGACGAAACCA----GAGGA------------- -----(((((((((.....))))-.........(((((((((.((....))...)))))))))))))).......--................----.....------------- ( -21.50) >DroMoj_CAF1 63045 101 + 1 AUUAAAUCUCUGGCAUUUAGCCA-G--AAUUUAUAAAUCAGUUGAACAAUUUGUGCUGAUUUAGUGACAUGAUAC--GA--A--U-GGAGCCAACUAAAGGA----CAGUAGCCA ..(((((.((((((.....))))-)--))))))(((((((((...((.....)))))))))))((..(((.....--..--)--)-)..))........((.----(....))). ( -22.70) >DroAna_CAF1 38779 90 + 1 AUUAUCGGGCUGGCACUUGGCCAGAAAAAUUUAUAAAUCAGUUGAGCAAUUUGUGCUGAUUUAGUGACAUGAUAC--CA--CAUUCGAAGCCA----G----------------- .......(((((((.....))).(((.......(((((((((.((....))...)))))))))(((.........--))--).)))..)))).----.----------------- ( -22.60) >consensus AUUAUAUCGCUGGCACUUGGCCA_A__AAUUUAUAAAUCAGUUGAACAAUUUGUGCUGAUUUAGUGACAUGAUAC__CA__A__U_GAAGCCA____AAGGA_____________ ......((((((((.....))))..........(((((((((...((.....)))))))))))))))................................................ (-13.50 = -13.92 + 0.42)
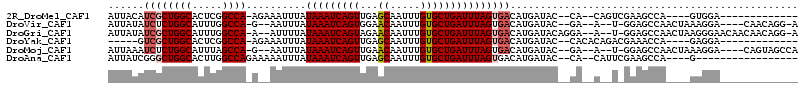
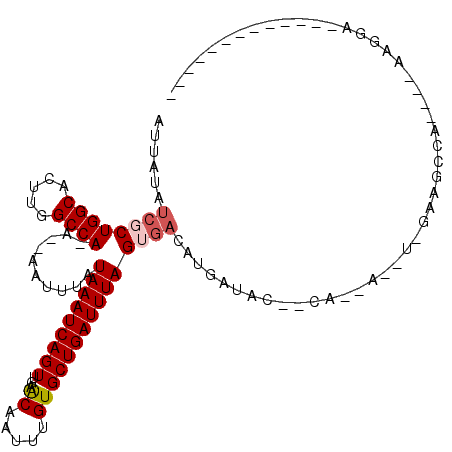
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:07 2006