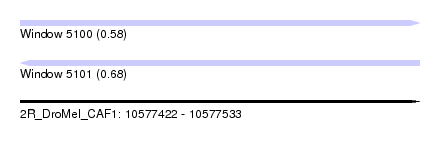
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,577,422 – 10,577,533 |
| Length | 111 |
| Max. P | 0.684382 |

| Location | 10,577,422 – 10,577,533 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -25.25 |
| Consensus MFE | -13.10 |
| Energy contribution | -13.30 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
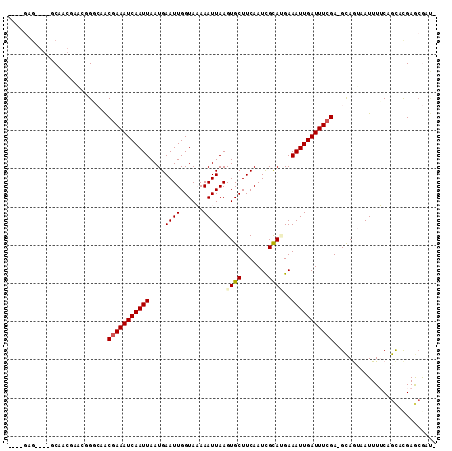
>2R_DroMel_CAF1 10577422 111 + 20766785 GCGUCACGGGAGCAACGAACGGCCAACGAAAUCAAUUAAUGAAUUGGUAAAAAUUAAGUGCUUCAAUCGCAUGAAAUUGAUUUCGA-GCAGUUAUUUUCAGCACGAACGAUG .((((...((((((.....((.....))..(((((((....)))))))..........))))))....((.(((((.(((((....-..))))).)))))))......)))) ( -22.80) >DroVir_CAF1 59418 100 + 1 ----GAG-----CAG--CAGCGGCAGCGAAAUCAAUUAAUGAAUUGGUAAAAAUUAAGUGCUUCAAUUGUAUGAAAUUGAUUUCGA-GUAAUAAUUUUCAGCUUGAGCGCCG ----..(-----(..--..))(((..(((((((((((.(((((((((...............)))))).)))..))))))))))).-.............((....))))). ( -22.86) >DroPse_CAF1 56236 106 + 1 ----GAGUGGUGCAACGAAUGGCGAACGAAAUCAAUUAAUGAAUUGGUACAAAUUAAGUGCUUCAAUCGCAAGAAAUUGAUUUCGA-GCAGAAAUUUGCAGCACGAGCAAU- ----..(((.(((((......((...(((((((((((..(((...(((((.......)))))))).((....))))))))))))).-))......))))).))).......- ( -33.10) >DroGri_CAF1 53032 100 + 1 ----GAG-----CGG--UGGUGGCAGCAAAAUCAAUUAAUGAAUUGGUAAAAAUUAAGUGCUUCAAUUGUACGAAAUUGAUUUCGA-GUAAUAAUUUUCAGUUCGAGCGACG ----..(-----(((--(..((....))..)))......((((((((.....((((..((..((((((......))))))...)).-.)))).....)))))))).)).... ( -18.10) >DroWil_CAF1 55156 107 + 1 GGAUCAGG--GACUUGGUGCAGGCAACGAAAUCAAUUAAUGAAUUGGUAAAAAUUAAGUGCUUCAAUUGCAUGAAAUUGAUUUCGAGGCAGUAAUUUUCAGCUCGAGAG--- (.(((((.--...))))).)..((..(((((((((((.(((((((((...............)))))).)))..)))))))))))..))......((((.....)))).--- ( -24.66) >DroPer_CAF1 57139 106 + 1 ----GAGUGGUGCAACGAAUGGAGAACGAAAUCAAUUAAUGAAUUGGUACAAAUUAAGUGCUUCAAUCGCAAGAAAUUGAUUUCGA-GCAGAAAUUUGCAGCACGAGCAAU- ----..((.((((.............(((((((((((..(((...(((((.......)))))))).((....))))))))))))).-((((....)))).))))..))...- ( -30.00) >consensus ____GAG____GCAACGAACGGGCAACGAAAUCAAUUAAUGAAUUGGUAAAAAUUAAGUGCUUCAAUCGCAUGAAAUUGAUUUCGA_GCAGUAAUUUUCAGCACGAGCGAU_ ..........................(((((((((((....((((......))))..((((.......))))..)))))))))))........................... (-13.10 = -13.30 + 0.20)

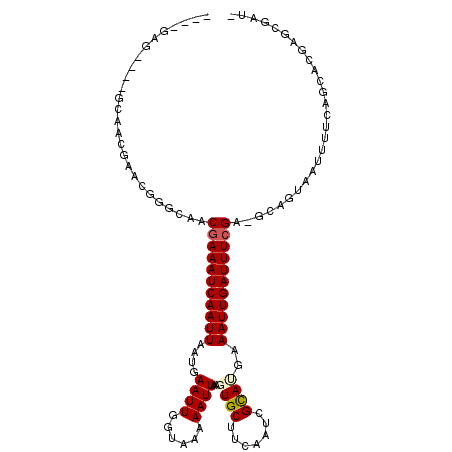
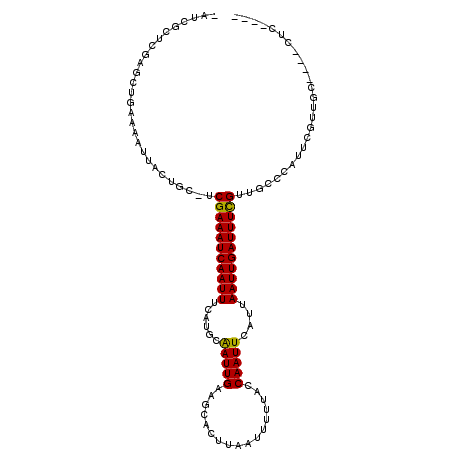
| Location | 10,577,422 – 10,577,533 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -20.30 |
| Consensus MFE | -11.20 |
| Energy contribution | -10.81 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10577422 111 - 20766785 CAUCGUUCGUGCUGAAAAUAACUGC-UCGAAAUCAAUUUCAUGCGAUUGAAGCACUUAAUUUUUACCAAUUCAUUAAUUGAUUUCGUUGGCCGUUCGUUGCUCCCGUGACGC ....(..((.((..(..........-.(((((((((((..(((.(((((.................)))))))).))))))))))))..))))..)((..(....)..)).. ( -22.33) >DroVir_CAF1 59418 100 - 1 CGGCGCUCAAGCUGAAAAUUAUUAC-UCGAAAUCAAUUUCAUACAAUUGAAGCACUUAAUUUUUACCAAUUCAUUAAUUGAUUUCGCUGCCGCUG--CUG-----CUC---- .(((((....)).............-.(((((((((((......(((((.................)))))....)))))))))))..)))((..--..)-----)..---- ( -18.93) >DroPse_CAF1 56236 106 - 1 -AUUGCUCGUGCUGCAAAUUUCUGC-UCGAAAUCAAUUUCUUGCGAUUGAAGCACUUAAUUUGUACCAAUUCAUUAAUUGAUUUCGUUCGCCAUUCGUUGCACCACUC---- -.......(((.(((((......((-.(((((((((((...((.(((((..(((.......)))..)))))))..)))))))))))...))......))))).)))..---- ( -27.40) >DroGri_CAF1 53032 100 - 1 CGUCGCUCGAACUGAAAAUUAUUAC-UCGAAAUCAAUUUCGUACAAUUGAAGCACUUAAUUUUUACCAAUUCAUUAAUUGAUUUUGCUGCCACCA--CCG-----CUC---- .((.((((.....))..........-.(((((((((((......(((((.................)))))....)))))))))))..)).))..--...-----...---- ( -9.23) >DroWil_CAF1 55156 107 - 1 ---CUCUCGAGCUGAAAAUUACUGCCUCGAAAUCAAUUUCAUGCAAUUGAAGCACUUAAUUUUUACCAAUUCAUUAAUUGAUUUCGUUGCCUGCACCAAGUC--CCUGAUCC ---.((..(..((..........((..(((((((((((..(((.(((((.................)))))))).)))))))))))..))........))..--)..))... ( -18.70) >DroPer_CAF1 57139 106 - 1 -AUUGCUCGUGCUGCAAAUUUCUGC-UCGAAAUCAAUUUCUUGCGAUUGAAGCACUUAAUUUGUACCAAUUCAUUAAUUGAUUUCGUUCUCCAUUCGUUGCACCACUC---- -.......(((.(((((......(.-.(((((((((((...((.(((((..(((.......)))..)))))))..)))))))))))..)........))))).)))..---- ( -25.24) >consensus _AUCGCUCGAGCUGAAAAUUACUGC_UCGAAAUCAAUUUCAUGCAAUUGAAGCACUUAAUUUUUACCAAUUCAUUAAUUGAUUUCGUUGCCCAUUCGUUGC____CUC____ ...........................(((((((((((......(((((.................)))))....))))))))))).......................... (-11.20 = -10.81 + -0.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:16 2006