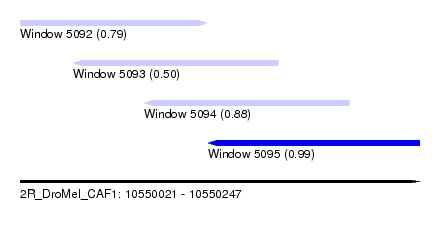
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,550,021 – 10,550,247 |
| Length | 226 |
| Max. P | 0.993374 |

| Location | 10,550,021 – 10,550,127 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.37 |
| Mean single sequence MFE | -18.37 |
| Consensus MFE | -17.19 |
| Energy contribution | -16.97 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.73 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10550021 106 + 20766785 AGUUGUCAAUGCUUUUAUAUACCUAUGUAGAUUGCCCUAAUGCUGAUUAGGUCGACGACAUAUAAACACACACUCAUGGGCAUAAAAUAUACAAAUGUACAUAUG--U .((((((...((.(((((((....)))))))..))((((((....))))))..))))))............((..(((..(((...........)))..)))..)--) ( -20.30) >DroSim_CAF1 39604 94 + 1 AGUUGUCAAUGCUUUUAUAUACCUAUGUAGAUUGCCCUAAUGCUGAUUAGGUCGACGACAUAUAGACACACACUCAUGGGCAUAAAAUAUAC--------------AU .((((((...((.(((((((....)))))))..))((((((....))))))..)))))).................................--------------.. ( -17.30) >DroEre_CAF1 44269 94 + 1 AGUUGUCAAUGCUUUUAUGUAGCUAUGUAGAUUGCCCUAAUGCUGAUUAGGUCGACGACAUAUAGACAAACACACAUGCUCGUA---UAUACA-----------AUAC .((((((...((.(((((((....)))))))..))((((((....))))))..))))))(((((((((........)).)).))---)))...-----------.... ( -17.50) >consensus AGUUGUCAAUGCUUUUAUAUACCUAUGUAGAUUGCCCUAAUGCUGAUUAGGUCGACGACAUAUAGACACACACUCAUGGGCAUAAAAUAUACA_____________AU .((((((...((.(((((((....)))))))..))((((((....))))))..))))))................................................. (-17.19 = -16.97 + -0.22)

| Location | 10,550,051 – 10,550,167 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.75 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -20.25 |
| Energy contribution | -20.48 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10550051 116 - 20766785 UUCGAUACCAAAUUGAGCGGCCUACUCACUCGGGGUAUACA--CAUAUGUACAUUUGUAUAUUUUAUGCCCAUGAGUGUGUGUUUAUAUGUCGUCGACCUAAUCAGCAUUAGGGCAAU .(((((..((...(((((.....((.(((((((((((((..--...((((((....))))))..))))))).)))))).)))))))..))..)))))((((((....))))))..... ( -35.20) >DroSim_CAF1 39634 104 - 1 UUCGAUACCAAAUUGAGCGGCCUACUCAUUUGGGGUAUAUAU--------------GUAUAUUUUAUGCCCAUGAGUGUGUGUCUAUAUGUCGUCGACCUAAUCAGCAUUAGGGCAAU .(((((..((...((..((.(.(((((((...(((((((...--------------........)))))))))))))).)))..))..))..)))))((((((....))))))..... ( -27.40) >DroEre_CAF1 44299 104 - 1 UUCGAUACCAAAUUGAGCGGCCUACUCACACGGGGUAUACGUAU-----------UGUAUA---UACGAGCAUGUGUGUUUGUCUAUAUGUCGUCGACCUAAUCAGCAUUAGGGCAAU .(((((..(.......(((...(((((.....)))))..)))..-----------.(((((---.(((((((....))))))).))))))..)))))((((((....))))))..... ( -25.30) >consensus UUCGAUACCAAAUUGAGCGGCCUACUCACUCGGGGUAUACAU_____________UGUAUAUUUUAUGCCCAUGAGUGUGUGUCUAUAUGUCGUCGACCUAAUCAGCAUUAGGGCAAU .(((((..((.((.((......(((.(((((((((((((.........................))))))).)))))).))))).)).))..)))))((((((....))))))..... (-20.25 = -20.48 + 0.23)

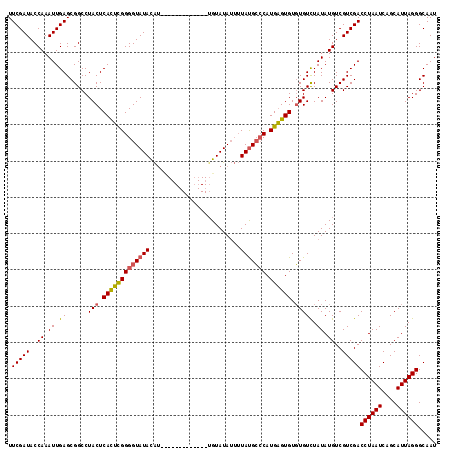
| Location | 10,550,091 – 10,550,207 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.75 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -22.73 |
| Energy contribution | -22.84 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10550091 116 - 20766785 AUUAGCAUUAAAAUGUUUUAGGAAAAACGACCGGGCCACGUUCGAUACCAAAUUGAGCGGCCUACUCACUCGGGGUAUACA--CAUAUGUACAUUUGUAUAUUUUAUGCCCAUGAGUG .............((((((....))))))...(((((..(((((((.....))))))))))))...(((((((((((((..--...((((((....))))))..))))))).)))))) ( -35.70) >DroSim_CAF1 39674 104 - 1 AUUAGCAUUAAAAUGUCUUAGGAAAAACGACCGGGCCACGUUCGAUACCAAAUUGAGCGGCCUACUCAUUUGGGGUAUAUAU--------------GUAUAUUUUAUGCCCAUGAGUG ....((((....))))....((........))(((((..(((((((.....))))))))))))((((((...(((((((...--------------........))))))))))))). ( -28.60) >DroEre_CAF1 44339 104 - 1 AUUAGCAUUAAAAUGUCUUAGGAAAAACGAGCGGGCCACGUUCGAUACCAAAUUGAGCGGCCUACUCACACGGGGUAUACGUAU-----------UGUAUA---UACGAGCAUGUGUG ....((((....))))............(((..((((..(((((((.....)))))))))))..)))(((((..((...(((((-----------.....)---)))).)).))))). ( -28.30) >consensus AUUAGCAUUAAAAUGUCUUAGGAAAAACGACCGGGCCACGUUCGAUACCAAAUUGAGCGGCCUACUCACUCGGGGUAUACAU_____________UGUAUAUUUUAUGCCCAUGAGUG ....((((....))))................(((((..(((((((.....))))))))))))...(((((((((((((.........................))))))).)))))) (-22.73 = -22.84 + 0.12)

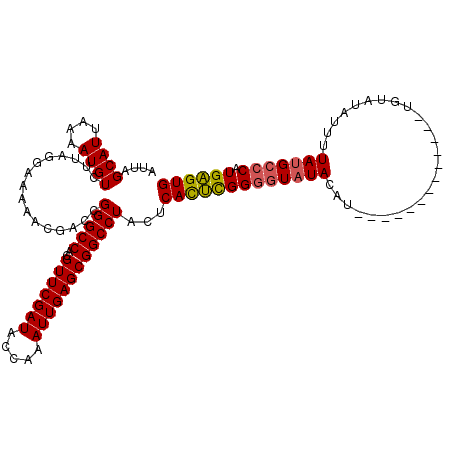
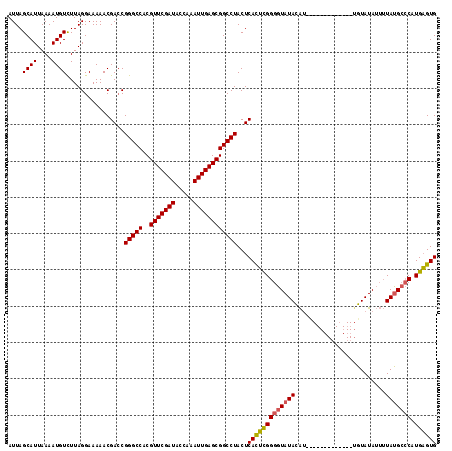
| Location | 10,550,127 – 10,550,247 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -36.13 |
| Consensus MFE | -33.26 |
| Energy contribution | -33.10 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10550127 120 - 20766785 GCCCACACCUAGGCUAAAGUAGGUCUACAGCUUGCGGCUCAUUAGCAUUAAAAUGUUUUAGGAAAAACGACCGGGCCACGUUCGAUACCAAAUUGAGCGGCCUACUCACUCGGGGUAUAC ((((.....(((((....((((((.....))))))(((((...(((((....)))))...((........))))))).((((((((.....)))))))))))))........)))).... ( -36.52) >DroSec_CAF1 47915 120 - 1 GCCCACACCUAGGCUAAAGUAGGUCUACAGCUUGCGGCUCAUUAGCAUUAAAAUGUCUUAGGAAAAACGACCGGGCCACGUUCGAUACCAAAUUGAGCGGCCUACUCACUCGGUAUAUAU (((......(((((....((((((.....))))))(((((....((((....))))....((........))))))).((((((((.....))))))))))))).......)))...... ( -32.62) >DroSim_CAF1 39698 120 - 1 GCCCACACCUAGGCUAAAGUAGGUCUACAGCUUGCGGCUCAUUAGCAUUAAAAUGUCUUAGGAAAAACGACCGGGCCACGUUCGAUACCAAAUUGAGCGGCCUACUCAUUUGGGGUAUAU ((((.....(((((....((((((.....))))))(((((....((((....))))....((........))))))).((((((((.....)))))))))))))........)))).... ( -36.02) >DroEre_CAF1 44363 120 - 1 GCCCACACCUAGGCCAAAGUAGGUCUACAGCUUGCGGCUCAUUAGCAUUAAAAUGUCUUAGGAAAAACGAGCGGGCCACGUUCGAUACCAAAUUGAGCGGCCUACUCACACGGGGUAUAC ((((...((((((.((..((((((.....)))))).(((....))).......)).))))))......(((..((((..(((((((.....)))))))))))..))).....)))).... ( -41.40) >DroYak_CAF1 44536 120 - 1 GCCCACACCUAGGCCAAAGUAGGUCUACAGCUUGCGGCUCAUUAGCAUUAAAAUGUCUUAGGAAAAACGACCGGGCCACGUUCGAUACCAAAUUGAACAGCCUACUCACUCGGGGUAUAU ((((...((((((.((..((((((.....)))))).(((....))).......)).))))))......((..((((...(((((((.....))))))).))))..)).....)))).... ( -34.10) >consensus GCCCACACCUAGGCUAAAGUAGGUCUACAGCUUGCGGCUCAUUAGCAUUAAAAUGUCUUAGGAAAAACGACCGGGCCACGUUCGAUACCAAAUUGAGCGGCCUACUCACUCGGGGUAUAU .(((...((((((.((..((((((.....)))))).(((....))).......)).))))))......((..(((((..(((((((.....))))))))))))..))....)))...... (-33.26 = -33.10 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:11 2006