| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,549,826 – 10,549,981 |
| Length | 155 |
| Max. P | 0.694832 |

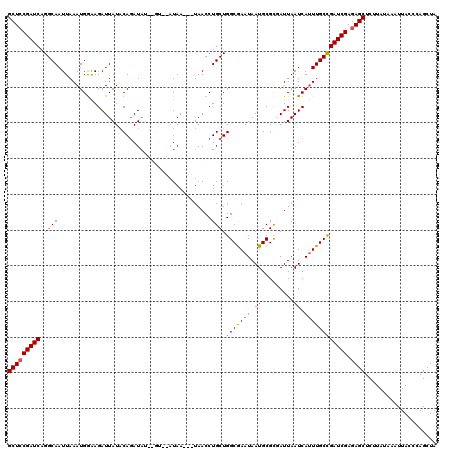
| Location | 10,549,826 – 10,549,941 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.08 |
| Mean single sequence MFE | -27.23 |
| Consensus MFE | -20.39 |
| Energy contribution | -20.39 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
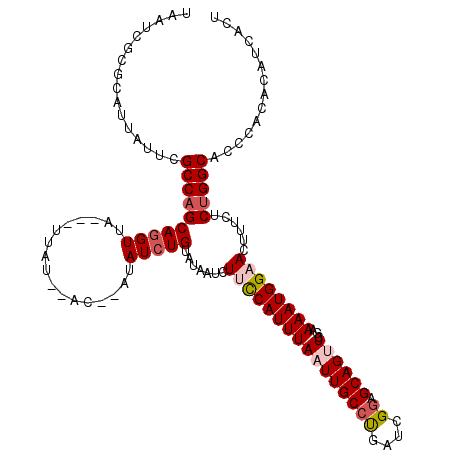
>2R_DroMel_CAF1 10549826 115 - 20766785 GCUCCGAUCAGGCAAUUAAAUGGAAGAUUAUUCAUAUAUAUGU--AUAA---UAACCUGCUGGCGAAUAAUGCGUGAUUAAUCAUUUGCCGAUCGAGAGCUCUUAUAAAUUACCCAGCUA (((((((((.((((.......((...(((((.(((....))).--))))---)..))))))(((((((.((.........)).)))))))))))).)))).................... ( -27.61) >DroSec_CAF1 47614 111 - 1 GCUCCGAUCAGGCAAUUAAAUGGAAGAUUAUACAGAUAU------AUAA---UAACCUGCUGGCGAAUAAUGCGCUAUUAAUCAUUUGCCGAUCGAGAGCUCCUAUAAAUUACCCAGCUA (((((((((.(((((......((...((((((......)------))))---)..))...(((((.......)))))........)))))))))).)))).................... ( -28.40) >DroSim_CAF1 39413 111 - 1 GCUCCGAUCAGGCAAUUAAAUGAAAGAUUAUACAGAUAU------UUAA---UAACCUGCUGGCGAAUAAUGCGCGAUUAAUCAAUUGCCGAUCGAGAGCUCUUAUAAAUUACCCAGCUA (((((((((.(((((((........(((((..(((....------....---.......)))(((.......)))...))))))))))))))))).)))).................... ( -26.66) >DroEre_CAF1 44073 116 - 1 GCUGCGAUCGGGCAAUUAAAUGGGAGAUUAUACAGAUGGGAGUAUAUAA---UAACCUGCUGGCAAAUAAUGCGCGAUUAAUCAUUUGCCGAUCGAGAGCUCUUAUAAAUUAC-CAGCUA (((.(((((((.....(((((((..((((...(((.((((.........---...)))))))(((.....)))..))))..))))))))))))))..))).............-...... ( -26.30) >DroYak_CAF1 44229 120 - 1 GCUCCGAUCGGGCAAUUAAAUGGGAGAUUAUACAGAUACGAGUAUAUAAUAAUAACCUGCUGGCAAAUAAUGCGCCAUUAAUCAUUUGCUGAUCGAGAGCUCUUAUAAAUUACCCAGCUA (((((((((((.(((......(((..(((((....(((....)))...)))))..)))..((((.........))))........)))))))))).)))).................... ( -27.20) >consensus GCUCCGAUCAGGCAAUUAAAUGGAAGAUUAUACAGAUAU__GU__AUAA___UAACCUGCUGGCGAAUAAUGCGCGAUUAAUCAUUUGCCGAUCGAGAGCUCUUAUAAAUUACCCAGCUA (((((((((.((((...........................................))))(((((((.((.........)).)))))))))))).)))).................... (-20.39 = -20.39 + 0.00)


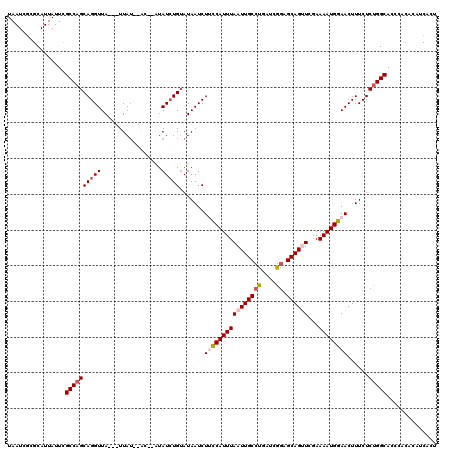
| Location | 10,549,866 – 10,549,981 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.05 |
| Mean single sequence MFE | -23.84 |
| Consensus MFE | -17.47 |
| Energy contribution | -18.47 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10549866 115 + 20766785 UAAUCACGCAUUAUUCGCCAGCAGGUUA---UUAU--ACAUAUAUAUGAAUAAUCUUCCAUUUAAUUGCCUGAUCGGAGCAGUUCGAAAAUGGAACUUUCUCUGGCACCCACACAUCACU ................(((((.((((((---((((--(......)))).)))))))(((((((((((((((....)).))))))...))))))).......))))).............. ( -25.20) >DroSec_CAF1 47654 111 + 1 UAAUAGCGCAUUAUUCGCCAGCAGGUUA---UUAU------AUAUCUGUAUAAUCUUCCAUUUAAUUGCCUGAUCGGAGCAGUUCGAAAAUGGAACUUUCUCUGGCACCCACACAUCACU ................(((((.(((..(---((((------((....))))))).((((((((((((((((....)).))))))...))))))))...)))))))).............. ( -25.70) >DroSim_CAF1 39453 111 + 1 UAAUCGCGCAUUAUUCGCCAGCAGGUUA---UUAA------AUAUCUGUAUAAUCUUUCAUUUAAUUGCCUGAUCGGAGCAGUUCGAAAAUGGAACUUUCUCUGGCACCCACACAUCACC ................((((((((((.(---((((------((....(......)....))))))).)))))...((((.(((((.......)))))))))))))).............. ( -24.20) >DroEre_CAF1 44112 117 + 1 UAAUCGCGCAUUAUUUGCCAGCAGGUUA---UUAUAUACUCCCAUCUGUAUAAUCUCCCAUUUAAUUGCCCGAUCGCAGCAGCUCCAAAAUGGAACUUUCUCCGGCACCCACACAACACU ...............(((((((.((..(---(((((((........))))))))..)).......((((......))))..))).......(((......)))))))............. ( -19.70) >DroYak_CAF1 44269 120 + 1 UAAUGGCGCAUUAUUUGCCAGCAGGUUAUUAUUAUAUACUCGUAUCUGUAUAAUCUCCCAUUUAAUUGCCCGAUCGGAGCAGCUCGAAAAUGGAACUUUCUCUGGCACCCACACAUCAUC ...............((((((.(((((.......(((((........))))).....((((((..((((((....)).)))).....)))))))))))...))))))............. ( -24.40) >consensus UAAUCGCGCAUUAUUCGCCAGCAGGUUA___UUAU__AC__AUAUCUGUAUAAUCUUCCAUUUAAUUGCCUGAUCGGAGCAGUUCGAAAAUGGAACUUUCUCUGGCACCCACACAUCACU ................((((((((((.................))))).......((((((((((((((((....)).))))))...))))))))......))))).............. (-17.47 = -18.47 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:07 2006