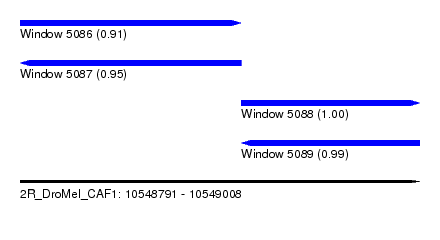
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,548,791 – 10,549,008 |
| Length | 217 |
| Max. P | 0.996974 |

| Location | 10,548,791 – 10,548,911 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.92 |
| Mean single sequence MFE | -29.41 |
| Consensus MFE | -26.64 |
| Energy contribution | -27.48 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10548791 120 + 20766785 GGAAACCCAUGAAUGAAAUGCUAAACCGGUUGGGCCUUUCAGCCUGCUGCUCAUCUUUCAUUUACUUUGCACUUACUUCACUCAGUCGAGUGUGUGUUUUGUAGUCUUUCCUUCUUUUAG (((((..(.(((((((((........((((.((((......))))))))......)))))))))...((((..(((..(((((....))))).)))...)))))..)))))......... ( -27.84) >DroSec_CAF1 46594 120 + 1 GGAAACCCAUGAAUGAAAUGCUAAACCGGUUGGGCCUUUCCGCCUGCUGCUCAUCUUUCAUUUACUUUGCACUUACUUCACUCAGUCGAGUGUGUGUUUUGUAGUCUUUCCUUCUUUUAG (((((..(.(((((((((........((((.((((......))))))))......)))))))))...((((..(((..(((((....))))).)))...)))))..)))))......... ( -27.74) >DroSim_CAF1 38392 120 + 1 GGAAACCCAUGAAUGAAAUGCUAAACCGGUUGGGCCUUUCGGCCUGCUGCUCAUCUUUCAUUUACUUUGCACUUACUUCACCCAGUCGAGUGUGUGUUUUGUAGUCUUUCCUUCUUUUAG (((((..(..((((((((........((((.(((((....)))))))))......))))))))((...((((.(((((.((...)).))))).))))...)).)..)))))......... ( -29.04) >DroEre_CAF1 43054 114 + 1 GGAAACCCAUGAAUGAAAUGCUAAACCGGUUGGGCCUUUCGGCCUGCUGCUCAUCUUUCAUUUACUUUGCACUUACUUCACUCAGUCGAGUGUGUGUUUCGCAGUCUUUCCUUC------ (((((..(.(((((((((........((((.(((((....)))))))))......)))))))))...(((...(((..(((((....))))).)))....))))..)))))...------ ( -31.54) >DroYak_CAF1 43137 120 + 1 GGAAACCCAUGAAUGAAAUGCUAAACCGGUUGGGCCUUUCGGCCUGCUGCUCAUCUUUCAUUUACUUUGCACUUACUGCACUCAGUCGAGUGUGUGUUUCGUAGUCUUUCCUUCUUUUAG (((((..(....(((((((((.....((((.(((((....)))))))))...................((((((((((....)))).))))))))))))))).)..)))))......... ( -30.90) >consensus GGAAACCCAUGAAUGAAAUGCUAAACCGGUUGGGCCUUUCGGCCUGCUGCUCAUCUUUCAUUUACUUUGCACUUACUUCACUCAGUCGAGUGUGUGUUUUGUAGUCUUUCCUUCUUUUAG (((((..(.(((((((((........((((.(((((....)))))))))......)))))))))...((((..(((..(((((....))))).)))...)))))..)))))......... (-26.64 = -27.48 + 0.84)

| Location | 10,548,791 – 10,548,911 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.92 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -28.10 |
| Energy contribution | -27.86 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10548791 120 - 20766785 CUAAAAGAAGGAAAGACUACAAAACACACACUCGACUGAGUGAAGUAAGUGCAAAGUAAAUGAAAGAUGAGCAGCAGGCUGAAAGGCCCAACCGGUUUAGCAUUUCAUUCAUGGGUUUCC .........(((((..((((........(((((....)))))..((....))...)))(((((((..(((((....((((....))))......)))))...)))))))...)..))))) ( -27.00) >DroSec_CAF1 46594 120 - 1 CUAAAAGAAGGAAAGACUACAAAACACACACUCGACUGAGUGAAGUAAGUGCAAAGUAAAUGAAAGAUGAGCAGCAGGCGGAAAGGCCCAACCGGUUUAGCAUUUCAUUCAUGGGUUUCC .........(((((..((((........(((((....)))))..((....))...)))(((((((..(((((....(((......)))......)))))...)))))))...)..))))) ( -23.90) >DroSim_CAF1 38392 120 - 1 CUAAAAGAAGGAAAGACUACAAAACACACACUCGACUGGGUGAAGUAAGUGCAAAGUAAAUGAAAGAUGAGCAGCAGGCCGAAAGGCCCAACCGGUUUAGCAUUUCAUUCAUGGGUUUCC .........(((((..((((........(((((....)))))..((....))...)))(((((((..(((((....((((....))))......)))))...)))))))...)..))))) ( -29.70) >DroEre_CAF1 43054 114 - 1 ------GAAGGAAAGACUGCGAAACACACACUCGACUGAGUGAAGUAAGUGCAAAGUAAAUGAAAGAUGAGCAGCAGGCCGAAAGGCCCAACCGGUUUAGCAUUUCAUUCAUGGGUUUCC ------...(((((..(((((.......(((((....))))).......))))..((.(((((((..(((((....((((....))))......)))))...))))))).)))..))))) ( -33.64) >DroYak_CAF1 43137 120 - 1 CUAAAAGAAGGAAAGACUACGAAACACACACUCGACUGAGUGCAGUAAGUGCAAAGUAAAUGAAAGAUGAGCAGCAGGCCGAAAGGCCCAACCGGUUUAGCAUUUCAUUCAUGGGUUUCC .........(((((..(((.(((.....((((..((((....)))).)))).........(((((..(((((....((((....))))......)))))...)))))))).))).))))) ( -31.50) >consensus CUAAAAGAAGGAAAGACUACAAAACACACACUCGACUGAGUGAAGUAAGUGCAAAGUAAAUGAAAGAUGAGCAGCAGGCCGAAAGGCCCAACCGGUUUAGCAUUUCAUUCAUGGGUUUCC .........(((((..((((........(((((....)))))..((....))...)))(((((((..(((((....((((....))))......)))))...)))))))...)..))))) (-28.10 = -27.86 + -0.24)

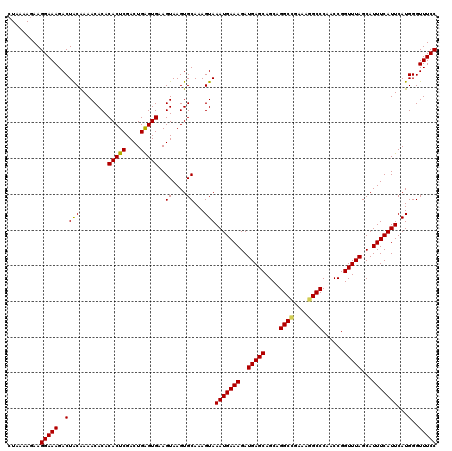
| Location | 10,548,911 – 10,549,008 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 85.64 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -20.67 |
| Energy contribution | -20.93 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.996974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10548911 97 + 20766785 UUUGCUGGCUGCUUCUUUGCUUCCUACUGUCACACUUCAAGGAUUUCUUUGUUGGCCAUCCUGGCCUGCAUAUGUGUGACAAUUACCCUCUGUA------UAU-- ..(((.((..((......))..))...((((((((..(((((....)))))..((((.....)))).......))))))))..........)))------...-- ( -27.20) >DroSec_CAF1 46714 97 + 1 UUUGCUGGCUGCUUCUUUGCUUCCUACUGUCACACUUCAAGGAUUUCUUUGUUGGCCAUCCUGGCCUCCAUAUGUGUGACAAUUACCCUCUGUA------UGU-- ..(((.((..((......))..))...((((((((..(((((....)))))..((((.....)))).......))))))))..........)))------...-- ( -27.20) >DroSim_CAF1 38512 97 + 1 UUUGCUGGCUGCUUCUUUGCUUCCUACUGUCACACUUCAAGGAUUUCUUUGUUGGCCAUCCUGGACUGCAUAUGUGUGACAAUUACCCUCUGUA------UAU-- ..(((.((..((......))..))...((((((((..(((((....)))))..((((.....)).))......))))))))..........)))------...-- ( -21.40) >DroEre_CAF1 43168 90 + 1 ---------------UUCGCUUCCUACUGUCACACUUCAAGGAUUUCUUUGUUGGCCAUCCUGGCCUGCAUAUGUGUGACAAUUACCCUCUAUACCUCUAUACCC ---------------............((((((((..(((((....)))))..((((.....)))).......))))))))........................ ( -22.60) >consensus UUUGCUGGCUGCUUCUUUGCUUCCUACUGUCACACUUCAAGGAUUUCUUUGUUGGCCAUCCUGGCCUGCAUAUGUGUGACAAUUACCCUCUGUA______UAU__ ...........................((((((((..(((((....)))))..((((.....)))).......))))))))........................ (-20.67 = -20.93 + 0.25)

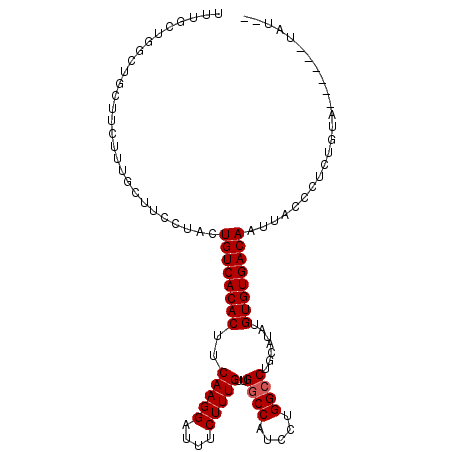
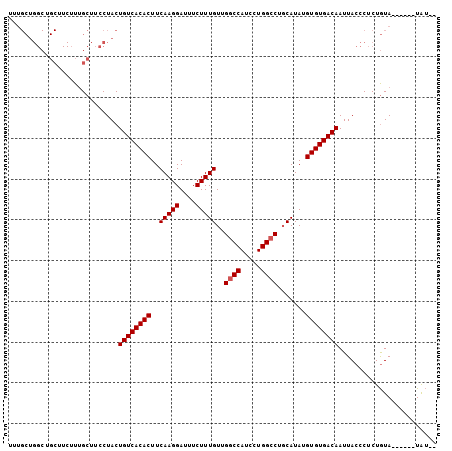
| Location | 10,548,911 – 10,549,008 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 85.64 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -21.27 |
| Energy contribution | -21.52 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10548911 97 - 20766785 --AUA------UACAGAGGGUAAUUGUCACACAUAUGCAGGCCAGGAUGGCCAACAAAGAAAUCCUUGAAGUGUGACAGUAGGAAGCAAAGAAGCAGCCAGCAAA --...------....(..(((.((((((((((.......((((.....))))..(((.(....).)))..)))))))))).....((......)).)))..)... ( -28.90) >DroSec_CAF1 46714 97 - 1 --ACA------UACAGAGGGUAAUUGUCACACAUAUGGAGGCCAGGAUGGCCAACAAAGAAAUCCUUGAAGUGUGACAGUAGGAAGCAAAGAAGCAGCCAGCAAA --...------....(..(((.((((((((((...((..((((.....))))..))(((.....)))...)))))))))).....((......)).)))..)... ( -29.40) >DroSim_CAF1 38512 97 - 1 --AUA------UACAGAGGGUAAUUGUCACACAUAUGCAGUCCAGGAUGGCCAACAAAGAAAUCCUUGAAGUGUGACAGUAGGAAGCAAAGAAGCAGCCAGCAAA --...------....(..(((.((((((((((........((.(((((..(.......)..))))).)).)))))))))).....((......)).)))..)... ( -24.10) >DroEre_CAF1 43168 90 - 1 GGGUAUAGAGGUAUAGAGGGUAAUUGUCACACAUAUGCAGGCCAGGAUGGCCAACAAAGAAAUCCUUGAAGUGUGACAGUAGGAAGCGAA--------------- ......................((((((((((.......((((.....))))..(((.(....).)))..))))))))))..........--------------- ( -23.20) >consensus __AUA______UACAGAGGGUAAUUGUCACACAUAUGCAGGCCAGGAUGGCCAACAAAGAAAUCCUUGAAGUGUGACAGUAGGAAGCAAAGAAGCAGCCAGCAAA ......................((((((((((.......((((.....))))..(((.(....).)))..))))))))))......................... (-21.27 = -21.52 + 0.25)

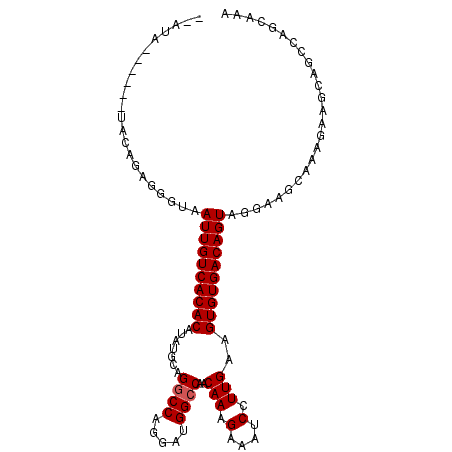
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:05 2006