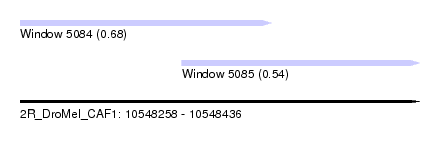
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,548,258 – 10,548,436 |
| Length | 178 |
| Max. P | 0.684641 |

| Location | 10,548,258 – 10,548,370 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.92 |
| Mean single sequence MFE | -22.92 |
| Consensus MFE | -12.73 |
| Energy contribution | -12.98 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
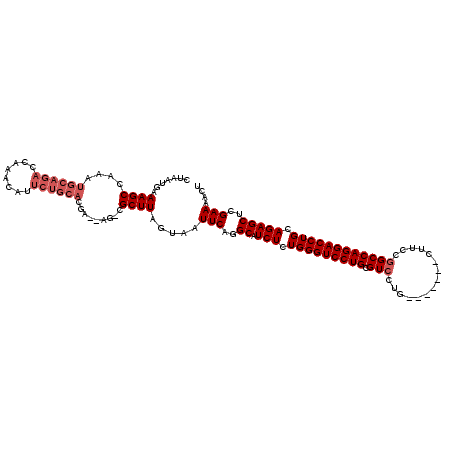
>2R_DroMel_CAF1 10548258 112 + 20766785 AAACAAAUACUUAACUCCUAGUUUUCAAACAGUAACUGA--------GCAGACAACUGCGAAAGUAAAAAUGAAGUGCGGCUAAUGAAAGCCAAAUGCAGCCCAAACAUUCUGCACGAAC .......(((((......(((((..........))))).--------((((....))))..)))))........((((((((......)))).((((.........))))..)))).... ( -20.20) >DroSec_CAF1 46079 90 + 1 AAACGA---------UCC---UUUUCGAUCAGUAACUCA--------GCAGAAAACUGCAAAAGUGAAAAUGGAGUGCGGCUAAUGAAAGCCAAAUGCAGACCAAACAUU---------- ....((---------((.---.....)))).(((((((.--------((((....))))....((....)).))))..((((......))))...)))............---------- ( -19.20) >DroSim_CAF1 37877 100 + 1 AAACGA---------UCC---UUUUCGAUCAGUAACUCA--------GCAGAAAACUGCAAAAGCGAAAAUGGAGUGCGGCUAAUGAAAGCCAAAUGCAGACCAAACAUUCUGCACGAGG ....((---------((.---.....)))).....(((.--------((((((...(((....)))....(((..(((((((......))))....)))..)))....))))))..))). ( -25.90) >DroYak_CAF1 42637 110 + 1 AAGCGAAUGCCCAACUCUUGCAUUUCGCACAGUAACUCAGUCACACGGCAGAGUCCUGCGAAAGGGAAAAUGAAGUGCGGCCAAUGAAAGCCAACUGCAGCUCAAAAGCC---------- ..((((((((.........))).))))).((((..........(((..((...((((......))))...))..))).(((........))).))))..(((....))).---------- ( -26.40) >consensus AAACGA_________UCC___UUUUCGAACAGUAACUCA________GCAGAAAACUGCAAAAGUGAAAAUGAAGUGCGGCUAAUGAAAGCCAAAUGCAGACCAAACAUU__________ ...............................................((((....))))...........(((..(((((((......))))....)))..)))................ (-12.73 = -12.98 + 0.25)



| Location | 10,548,330 – 10,548,436 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.29 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -23.68 |
| Energy contribution | -26.01 |
| Covariance contribution | 2.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10548330 106 + 20766785 CUAAUGAAAGCCAAAUGCAGCCCAAACAUUCUGCACGAACAGACGCUUAGUAAUUCAGGCAUCUCUGGGUCCUGCGU--------------CCGGCCAGGACCUGCAGAGCUCGAAAACU ....(((..(((...(((((..........))))).(((...((.....))..))).)))..((((((((((((.(.--------------....))))))))..))))).)))...... ( -29.50) >DroSec_CAF1 46139 107 + 1 CUAAUGAAAGCCAAAUGCAGACCAAACAUU-------------CGCUUAGUAAUUCAGGCAUCUCUGGGUCCUGCGUCCUGCUUCCUGCUUCCGGCCAGGACCUGCAGAGCUCGAAAACU .......((((..((((.........))))-------------.)))).....(((..((.(((.(((((((((.(((..((.....))....)))))))))))).)))))..))).... ( -30.40) >DroSim_CAF1 37937 113 + 1 CUAAUGAAAGCCAAAUGCAGACCAAACAUUCUGCACGAGGAGUCGCUUAGUAAUUCAGGCAUCUCUGGGUCCUGCGUCCUG-------CUUCCGGCCAGGACCUGCAGAGCUCGAAAACU (((((((...((...((((((........))))))...))..))).))))...(((..((.(((.(((((((((.(((...-------.....)))))))))))).)))))..))).... ( -38.30) >consensus CUAAUGAAAGCCAAAUGCAGACCAAACAUUCUGCACGA__AG_CGCUUAGUAAUUCAGGCAUCUCUGGGUCCUGCGUCCUG_______CUUCCGGCCAGGACCUGCAGAGCUCGAAAACU .......((((....((((((........)))))).........)))).....(((..((.(((.(((((((((.(((...............)))))))))))).)))))..))).... (-23.68 = -26.01 + 2.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:02 2006