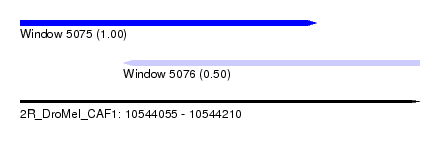
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,544,055 – 10,544,210 |
| Length | 155 |
| Max. P | 0.997646 |

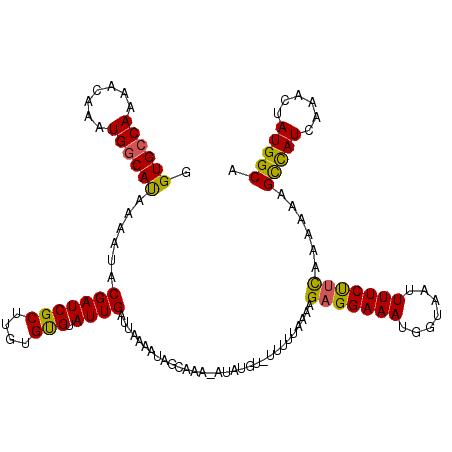
| Location | 10,544,055 – 10,544,170 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.52 |
| Mean single sequence MFE | -25.87 |
| Consensus MFE | -23.48 |
| Energy contribution | -22.60 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.997646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10544055 115 + 20766785 GGUGCCAAAACAAAUGGCACAAAAUACGAUCGCUUGUGUGUAUUGAUUAAAAUAGCAAA-AUA----UUUUUAAAAGAGGAAAUGGUAAUUUUCUUCAAAAAAGUCAUCAAACUAUGGCA .((((((.......))))))...........(((...((....(((((((((((.....-.))----)))).....(((((((.......))))))).....)))))....))...))). ( -25.10) >DroSec_CAF1 37175 119 + 1 GGUGCCAAAACAAAUGGCAUAAAAUACGAUCGCUUGUGUGUAUUGAUUUAAAUAGCAAA-AUAUGUAUUUUUAAAAGAGGAAAUGGUAAUUUUCUUCAAAAAAGCCAUCAAACUAUGGCA .((((((.......))))))......(((((((....))).)))).((((((..(((..-...)))...)))))).(((((((.......)))))))......(((((......))))). ( -26.20) >DroSim_CAF1 33736 119 + 1 GGUGCCAAAACAAAUGGCAUAAAAUACGAUCGCUUGUGCGUAUUGAUUAAAAUAGCAAA-AUAUGUAUUUUUAAAAGAGGAAAUGGUAAUUUUCUUCAAAAAAGCCAUCAAACUAUGGCA .((((((.......))))))......(((((((....))).)))).((((((..(((..-...)))..))))))..(((((((.......)))))))......(((((......))))). ( -28.40) >DroEre_CAF1 38564 117 + 1 GGUGGCAAAACAAAUGGCACAAAAUUCGAUCGCCUGUGCGAAUUGAUUAUAUUUGCAAA-AUAUUU--UUUGAAAAGAGGAAAUGCUAAUUUUCCCUAAAAAAGCCAUCAAACUAUGGCA ((((((....((((.(((.............)))..(((((((.......)))))))..-......--))))...((.(((((.......)))))))......))))))........... ( -24.92) >DroYak_CAF1 38436 120 + 1 GGUGCCAAAACAAAUGGCAUAAACUGCGAUCGCUGGUGUGUAUUGAUUGUAUUUGCAAAAAUAUUUUGUUUGAAAAGAGGAAAUGGUAAUUUUCCUUAAAAAAGCUAUCAAACUAUGGCG ...((((......(((((.(((((..(((((((....))).))))...((((((....))))))...)))))....(((((((.......)))))))......))))).......)))). ( -24.72) >consensus GGUGCCAAAACAAAUGGCAUAAAAUACGAUCGCUUGUGUGUAUUGAUUAAAAUAGCAAA_AUAUGU_UUUUUAAAAGAGGAAAUGGUAAUUUUCUUCAAAAAAGCCAUCAAACUAUGGCA .((((((.......))))))......(((((((....))).))))...............................(((((((.......)))))))......(((((......))))). (-23.48 = -22.60 + -0.88)

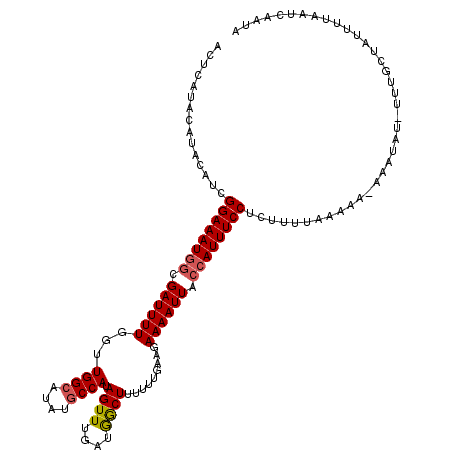
| Location | 10,544,095 – 10,544,210 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.80 |
| Mean single sequence MFE | -22.42 |
| Consensus MFE | -17.84 |
| Energy contribution | -18.12 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10544095 115 - 20766785 ACUCAUACAUACAUCGGAAAUGGAGAUUUUGGUUGGCAUAUGCCAUAGUUUGAUGACUUUUUUGAAGAAAAUUACCAUUUCCUCUUUUAAAAA----UAU-UUUGCUAUUUUAAUCAAUA ...............((((((((.((((((...((((....)))).((((....)))).........)))))).)))))))).......((((----((.-.....))))))........ ( -21.90) >DroSec_CAF1 37215 119 - 1 ACUCAUACAUACAUCGGAAAUGGCGAUUUUGGUUGGCAUAUGCCAUAGUUUGAUGGCUUUUUUGAAGAAAAUUACCAUUUCCUCUUUUAAAAAUACAUAU-UUUGCUAUUUAAAUCAAUA ...............((((((((..(((((..(..(.....(((((......)))))....)..)..)))))..))))))))........(((((((...-..)).)))))......... ( -25.10) >DroSim_CAF1 33776 119 - 1 ACUCAUACAUACAUCGGAAAUGGUGAUUUUGGUUGGCAUAUGCCAUAGUUUGAUGGCUUUUUUGAAGAAAAUUACCAUUUCCUCUUUUAAAAAUACAUAU-UUUGCUAUUUUAAUCAAUA ...............(((((((((((((((..(..(.....(((((......)))))....)..)..))))))))))))))).......((((((((...-..)).))))))........ ( -29.60) >DroEre_CAF1 38604 113 - 1 CCUACU----ACAUCGGAAAUGGCGAUUUUGGUUGGCAUAUGCCAUAGUUUGAUGGCUUUUUUAGGGAAAAUUAGCAUUUCCUCUUUUCAAA--AAAUAU-UUUGCAAAUAUAAUCAAUU ......----.....(((((((..((((((..((((.....(((((......)))))....))))..))))))..)))))))..(((.((((--(....)-)))).)))........... ( -18.80) >DroYak_CAF1 38476 116 - 1 AAUCAU----ACAUCGGAAAUUGCGAUUUUGGUUGGAAUACGCCAUAGUUUGAUAGCUUUUUUAAGGAAAAUUACCAUUUCCUCUUUUCAAACAAAAUAUUUUUGCAAAUACAAUCAAUA ......----..........((((((...((((........))))..((((((.((........((((((.......))))))))..)))))).........))))))............ ( -16.70) >consensus ACUCAUACAUACAUCGGAAAUGGCGAUUUUGGUUGGCAUAUGCCAUAGUUUGAUGGCUUUUUUGAAGAAAAUUACCAUUUCCUCUUUUAAAAA_AAAUAU_UUUGCUAUUUUAAUCAAUA ...............((((((((.((((((...((((....)))).((((....)))).........)))))).))))))))...................................... (-17.84 = -18.12 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:53 2006