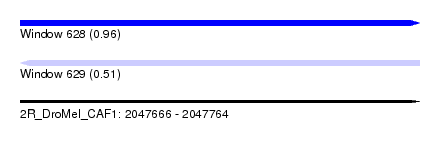
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,047,666 – 2,047,764 |
| Length | 98 |
| Max. P | 0.964728 |

| Location | 2,047,666 – 2,047,764 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 75.36 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -13.89 |
| Energy contribution | -15.45 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2047666 98 + 20766785 GUUUGCCAACCAGCCACACACACAUACUCGUGUAUACAUACAUAUGUGUCUAUAUAGUAUCUUUAUAUGGGAGUGCAGUAUAGAGAGUAAGGCAAACA (((((((.............((((((...(((....)))...))))))(((((((.(((((((.....)))).))).)))))))......))))))). ( -28.10) >DroSec_CAF1 32435 81 + 1 GUUUGCCAACCAGCCUGCCACACAUACCCAU----------------GUCUAUAUAGUAUCGUU-UAAGGGAGUGCAGUAUAGAGAGUAAGGCAAACA (((((((...............(((....))----------------)(((((((.(((((...-.....)).))).)))))))......))))))). ( -19.00) >DroSim_CAF1 33031 82 + 1 GUUUGCCAACCAGCCUGCCACACAUACUCAU----------------GUCUAUAUAGUAUCUUUAUAGGGGAGUGCAGUAUAGAGAGUAAGGCAAACA (((((((.....(....)......(((((..----------------.(((((((.(((((((.....)))).))).)))))))))))).))))))). ( -23.80) >DroYak_CAF1 35149 81 + 1 GUUUGCCAGCCAGCCA----CACAUAUAUGU--------AUGUAUGUAUCUAU-----AUCUGGAUAGGGGAGUGCAGUGCAGAGAGUAAGGCAAACA (((((((.((...((.----.(((((((...--------.))))))).(((((-----......)))))))...))..(((.....))).))))))). ( -22.60) >consensus GUUUGCCAACCAGCCAGCCACACAUACUCAU________________GUCUAUAUAGUAUCUUUAUAGGGGAGUGCAGUAUAGAGAGUAAGGCAAACA (((((((.....(.......)...........................(((((((.(((((((.....)))).))).)))))))......))))))). (-13.89 = -15.45 + 1.56)
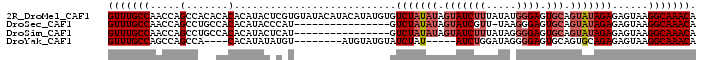
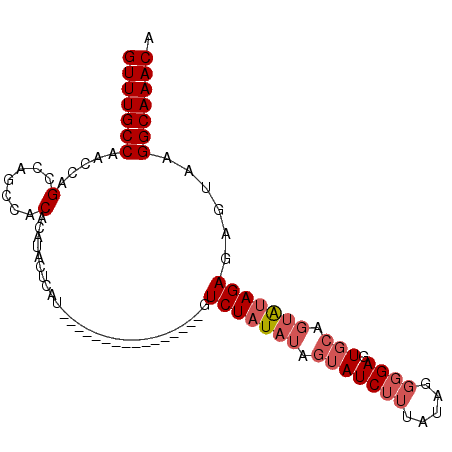
| Location | 2,047,666 – 2,047,764 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 75.36 |
| Mean single sequence MFE | -22.52 |
| Consensus MFE | -10.54 |
| Energy contribution | -11.79 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.47 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2047666 98 - 20766785 UGUUUGCCUUACUCUCUAUACUGCACUCCCAUAUAAAGAUACUAUAUAGACACAUAUGUAUGUAUACACGAGUAUGUGUGUGUGGCUGGUUGGCAAAC .(((((((..(((..((((((.((((............((((((((((......)))))).))))..........))))))))))..))).))))))) ( -27.05) >DroSec_CAF1 32435 81 - 1 UGUUUGCCUUACUCUCUAUACUGCACUCCCUUA-AACGAUACUAUAUAGAC----------------AUGGGUAUGUGUGGCAGGCUGGUUGGCAAAC .(((((((..(((..((...(..(((.(((...-.................----------------..)))...)))..)..))..))).))))))) ( -21.25) >DroSim_CAF1 33031 82 - 1 UGUUUGCCUUACUCUCUAUACUGCACUCCCCUAUAAAGAUACUAUAUAGAC----------------AUGAGUAUGUGUGGCAGGCUGGUUGGCAAAC .(((((((..(((((((((((...((((..(((((.((...)).)))))..----------------..))))..)))))).))...))).))))))) ( -21.70) >DroYak_CAF1 35149 81 - 1 UGUUUGCCUUACUCUCUGCACUGCACUCCCCUAUCCAGAU-----AUAGAUACAUACAU--------ACAUAUAUGUG----UGGCUGGCUGGCAAAC .(((((((.........((...((......((((......-----))))...(((((((--------(....))))))----))))..)).))))))) ( -20.10) >consensus UGUUUGCCUUACUCUCUAUACUGCACUCCCCUAUAAAGAUACUAUAUAGAC________________ACGAGUAUGUGUGGCAGGCUGGUUGGCAAAC .(((((((..(((..((...((((((.................................................))))))..))..))).))))))) (-10.54 = -11.79 + 1.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:04 2006