
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,540,293 – 10,540,687 |
| Length | 394 |
| Max. P | 0.999980 |

| Location | 10,540,293 – 10,540,392 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 93.19 |
| Mean single sequence MFE | -42.27 |
| Consensus MFE | -37.40 |
| Energy contribution | -37.40 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10540293 99 + 20766785 CGCUGGCGAACUGUUGACUGCCAGCUUGCUUGCCACUUCACGGUGAUG-CAGGGGAUGGCGGAUGGGUGGGUGG--------UGCUGGUGGUUGGUGGAUAUACAGCC .((((.....(..(..((..((((((..((..((...((...((.((.-(...).)).)).))..))..))..)--------.)))))..))..)..).....)))). ( -41.20) >DroSec_CAF1 33554 107 + 1 CGCUGGCGAACUGUUGACUGCCAGCUUGCUUGCCACUUCACGGUGAUG-UAGGGGAUGGCGGAUGGGUGGGUGGGUGGUUGGUGCUGGUGGUUGGUGGAUAUACAGCC .((((.....(..(..((..(((((..(((.((((((((((.....((-(........))).....))))...)))))).))))))))..))..)..).....)))). ( -41.00) >DroSim_CAF1 30004 108 + 1 CGCUGGCGAACUGUUGACUGCCAGCUUGCUUGCCACUUCACGGUGAUGGCAGGGGAUGGCGGAUGGGUGGGUGGUUGGUUGCUGCUGGUGGUUGGUGGAUAUACAGCC .((((.....(..(..((..(((((..(((.((((((.(((.....((.((.....)).)).....))))))))).)))....)))))..))..)..).....)))). ( -44.60) >consensus CGCUGGCGAACUGUUGACUGCCAGCUUGCUUGCCACUUCACGGUGAUG_CAGGGGAUGGCGGAUGGGUGGGUGG_UGGUUG_UGCUGGUGGUUGGUGGAUAUACAGCC .((((.....(..(..((..((((((..((..((...((...((.((........)).)).))..))..))..).........)))))..))..)..).....)))). (-37.40 = -37.40 + -0.00)

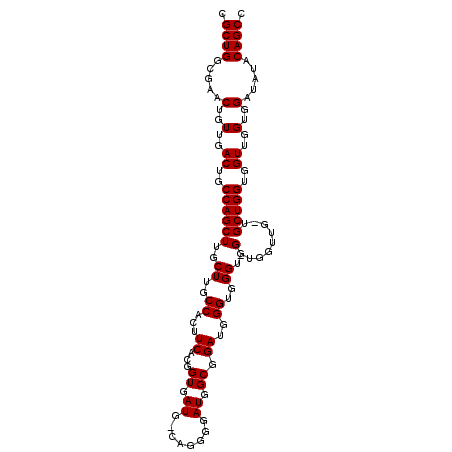
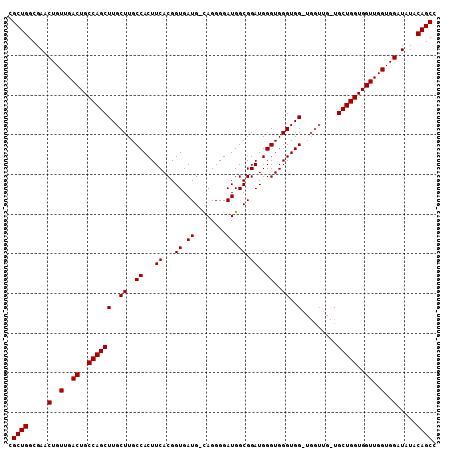
| Location | 10,540,360 – 10,540,472 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.80 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -26.71 |
| Energy contribution | -27.27 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10540360 112 + 20766785 GGGUGG--------UGCUGGUGGUUGGUGGAUAUACAGCCGGGUUUCCAUUGCAUUUUCGGUCAUUAUUCAACGUCAUUGCAGUUUGCUGCUGCGGUGGCAGCUUCAAUGAUGCAUAAUU ((((((--------((((((..(((((((((...((......)).))))))).))..)))).))))))))..((((((((.((((.((..(....)..)))))).))))))))....... ( -36.50) >DroSec_CAF1 33621 120 + 1 GGGUGGGUGGUUGGUGCUGGUGGUUGGUGGAUAUACAGCCGGGUUUCCAUUGCAUUUUCGGUCAUUAUUCAACGUCAUUGCAGUUUGCUGCUGCGGUGGCAGCUUCAAUGAUGCAUAAUU ..(((.((.(((((.(((.......(.((((((....((((((.............))))))...)))))).)(((((((((((.....)))))))))))))).))))).)).))).... ( -38.82) >DroSim_CAF1 30072 120 + 1 GGGUGGUUGGUUGCUGCUGGUGGUUGGUGGAUAUACAGCCGGGUUUCCAUUGCAUUUUCGGUCAUUAUUCAACGUCAUUGCAGUUUGCUGCUGCGGUGGCAGCUUCAAUGAUGCAUAAUU (.((.(((((..(((((........(.((((((....((((((.............))))))...)))))).)..(((((((((.....)))))))))))))).))))).)).)...... ( -37.12) >DroEre_CAF1 35047 93 + 1 GGGUGG--------UGCUGGUG-------------------GGUUUCCAUUGCAUUUUCGGUCAUUAUUCAACGUCAUUGCAGCUUGCUGCUGCGGUGGCAGCUUCAAUGAUGCAUAAUU .((..(--------(((.((((-------------------(....)))))))))..)).(.(((((((.((((((((((((((.....))))))))))).).)).))))))))...... ( -31.70) >consensus GGGUGG________UGCUGGUGGUUGGUGGAUAUACAGCCGGGUUUCCAUUGCAUUUUCGGUCAUUAUUCAACGUCAUUGCAGUUUGCUGCUGCGGUGGCAGCUUCAAUGAUGCAUAAUU ((((((........((((((((((((.........)))))).....)))..)))..........))))))..((((((((.((((.((..(....)..)))))).))))))))....... (-26.71 = -27.27 + 0.56)

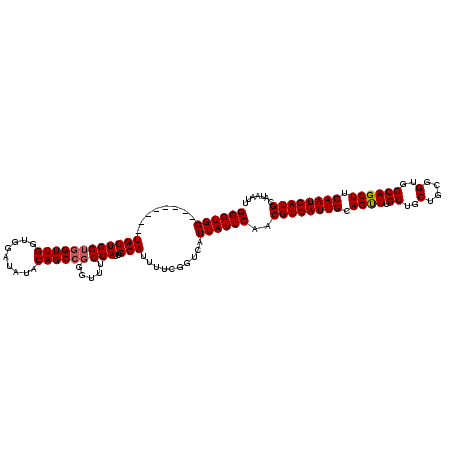
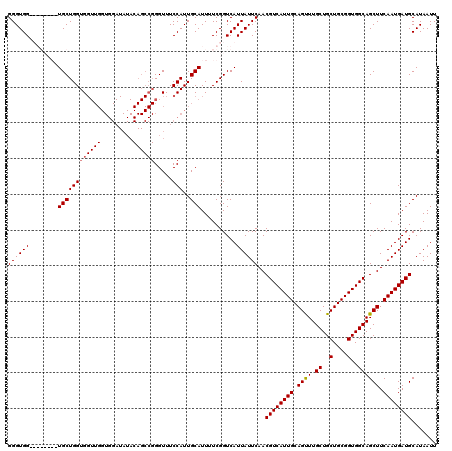
| Location | 10,540,392 – 10,540,512 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.33 |
| Mean single sequence MFE | -31.32 |
| Consensus MFE | -30.28 |
| Energy contribution | -30.12 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10540392 120 + 20766785 GGGUUUCCAUUGCAUUUUCGGUCAUUAUUCAACGUCAUUGCAGUUUGCUGCUGCGGUGGCAGCUUCAAUGAUGCAUAAUUUGUAGCGUUAUAUAAAUGCAAACUGUGUUGAUAUUUUUUA ((....)).........................((((..(((((((((.(((((....)))))....(((((((..........)))))))......)))))))))..))))........ ( -31.20) >DroSec_CAF1 33661 120 + 1 GGGUUUCCAUUGCAUUUUCGGUCAUUAUUCAACGUCAUUGCAGUUUGCUGCUGCGGUGGCAGCUUCAAUGAUGCAUAAUUUGUAGCGUUAUAUAAAUGCAAACUGUGUUGAUAUUUUUUA ((....)).........................((((..(((((((((.(((((....)))))....(((((((..........)))))))......)))))))))..))))........ ( -31.20) >DroSim_CAF1 30112 120 + 1 GGGUUUCCAUUGCAUUUUCGGUCAUUAUUCAACGUCAUUGCAGUUUGCUGCUGCGGUGGCAGCUUCAAUGAUGCAUAAUUUGUAGCGUUAUAUAAAUGCAAACUGUGUUGAUAUUUUUUA ((....)).........................((((..(((((((((.(((((....)))))....(((((((..........)))))))......)))))))))..))))........ ( -31.20) >DroEre_CAF1 35061 119 + 1 -GGUUUCCAUUGCAUUUUCGGUCAUUAUUCAACGUCAUUGCAGCUUGCUGCUGCGGUGGCAGCUUCAAUGAUGCAUAAUUUGUAGCGUUAUAUAAAUGCAAACUGUGUUGAUAUUUUUUA -.(((..((((((((((................(((((((((((.....))))))))))).......(((((((..........)))))))..)))))))....)))..)))........ ( -31.80) >DroYak_CAF1 34790 120 + 1 GGGUUUCCAUUGCAUUUUCGGUCAUUAUUCAACGUCAUUGCAGUUUGCUGCUGCGGUGGCAGCUUCAAUGAUGCAUAAUUUGUAGCGUUAUAUAAAUGCAAACUGUGUUGAUAUUUUUUA ((....)).........................((((..(((((((((.(((((....)))))....(((((((..........)))))))......)))))))))..))))........ ( -31.20) >consensus GGGUUUCCAUUGCAUUUUCGGUCAUUAUUCAACGUCAUUGCAGUUUGCUGCUGCGGUGGCAGCUUCAAUGAUGCAUAAUUUGUAGCGUUAUAUAAAUGCAAACUGUGUUGAUAUUUUUUA ..(((..((((((((((................(((((((((((.....))))))))))).......(((((((..........)))))))..)))))))....)))..)))........ (-30.28 = -30.12 + -0.16)

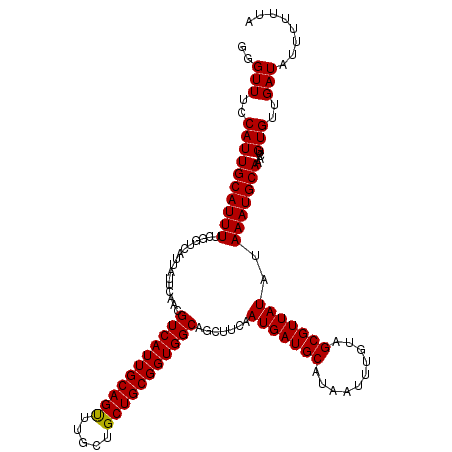
| Location | 10,540,392 – 10,540,512 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.33 |
| Mean single sequence MFE | -24.90 |
| Consensus MFE | -25.06 |
| Energy contribution | -24.90 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.43 |
| Structure conservation index | 1.01 |
| SVM decision value | 3.49 |
| SVM RNA-class probability | 0.999293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10540392 120 - 20766785 UAAAAAAUAUCAACACAGUUUGCAUUUAUAUAACGCUACAAAUUAUGCAUCAUUGAAGCUGCCACCGCAGCAGCAAACUGCAAUGACGUUGAAUAAUGACCGAAAAUGCAAUGGAAACCC .........(((...((((((((...........((..........)).........(((((....))))).))))))))...)))(((((....((........)).)))))....... ( -24.90) >DroSec_CAF1 33661 120 - 1 UAAAAAAUAUCAACACAGUUUGCAUUUAUAUAACGCUACAAAUUAUGCAUCAUUGAAGCUGCCACCGCAGCAGCAAACUGCAAUGACGUUGAAUAAUGACCGAAAAUGCAAUGGAAACCC .........(((...((((((((...........((..........)).........(((((....))))).))))))))...)))(((((....((........)).)))))....... ( -24.90) >DroSim_CAF1 30112 120 - 1 UAAAAAAUAUCAACACAGUUUGCAUUUAUAUAACGCUACAAAUUAUGCAUCAUUGAAGCUGCCACCGCAGCAGCAAACUGCAAUGACGUUGAAUAAUGACCGAAAAUGCAAUGGAAACCC .........(((...((((((((...........((..........)).........(((((....))))).))))))))...)))(((((....((........)).)))))....... ( -24.90) >DroEre_CAF1 35061 119 - 1 UAAAAAAUAUCAACACAGUUUGCAUUUAUAUAACGCUACAAAUUAUGCAUCAUUGAAGCUGCCACCGCAGCAGCAAGCUGCAAUGACGUUGAAUAAUGACCGAAAAUGCAAUGGAAACC- .........(((...((((((((...........((..........)).........(((((....))))).))))))))...)))(((((....((........)).)))))......- ( -24.90) >DroYak_CAF1 34790 120 - 1 UAAAAAAUAUCAACACAGUUUGCAUUUAUAUAACGCUACAAAUUAUGCAUCAUUGAAGCUGCCACCGCAGCAGCAAACUGCAAUGACGUUGAAUAAUGACCGAAAAUGCAAUGGAAACCC .........(((...((((((((...........((..........)).........(((((....))))).))))))))...)))(((((....((........)).)))))....... ( -24.90) >consensus UAAAAAAUAUCAACACAGUUUGCAUUUAUAUAACGCUACAAAUUAUGCAUCAUUGAAGCUGCCACCGCAGCAGCAAACUGCAAUGACGUUGAAUAAUGACCGAAAAUGCAAUGGAAACCC .........(((...((((((((...........((..........)).........(((((....))))).))))))))...)))(((((....((........)).)))))....... (-25.06 = -24.90 + -0.16)

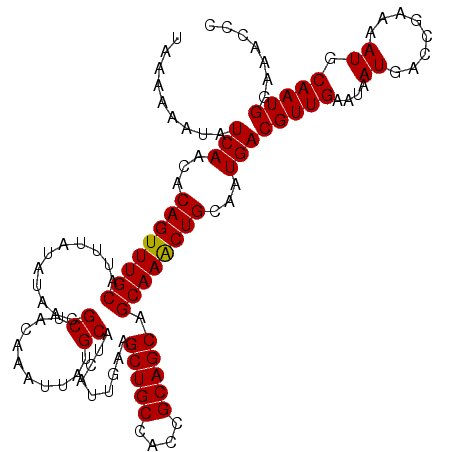

| Location | 10,540,432 – 10,540,548 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.49 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -29.08 |
| Energy contribution | -29.48 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.14 |
| SVM RNA-class probability | 0.999812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10540432 116 + 20766785 CAGUUUGCUGCUGCGGUGGCAGCUUCAAUGAUGCAUAAUUUGUAGCGUUAUAUAAAUGCAAACUGUGUUGAUAUUUUUUAUGCUAAUUGCAUGCAAGUAAAACUUACACACA----CACA .(((((((.(((((....)))))....(((((((..........)))))))......)))))))((((((........(((((.....)))))...(((.....))).)).)----))). ( -30.70) >DroSec_CAF1 33701 120 + 1 CAGUUUGCUGCUGCGGUGGCAGCUUCAAUGAUGCAUAAUUUGUAGCGUUAUAUAAAUGCAAACUGUGUUGAUAUUUUUUAUGCUAAUUGCAUGCAACUAAAACUUACACACACAUACACA ((((((((.(((((....)))))....(((((((..........)))))))......)))))))).((((........(((((.....)))))))))....................... ( -29.40) >DroSim_CAF1 30152 120 + 1 CAGUUUGCUGCUGCGGUGGCAGCUUCAAUGAUGCAUAAUUUGUAGCGUUAUAUAAAUGCAAACUGUGUUGAUAUUUUUUAUGCUAAUUGCAUGCAAGUAAAACUUACACACACAUACACA .(((((((.(((((....)))))....(((((((..........)))))))......)))))))((((((........(((((.....)))))...(((.....))).....)).)))). ( -30.70) >DroEre_CAF1 35100 118 + 1 CAGCUUGCUGCUGCGGUGGCAGCUUCAAUGAUGCAUAAUUUGUAGCGUUAUAUAAAUGCAAACUGUGUUGAUAUUUUUUAUGCUAAUUGCAUGCAAGUAAAACUUACAUACACUC--ACA ..((((((.(((((....))))).......(((((.......((((((.((((.(((((.....))))).)))).....))))))..))))))))))).................--... ( -29.20) >DroYak_CAF1 34830 118 + 1 CAGUUUGCUGCUGCGGUGGCAGCUUCAAUGAUGCAUAAUUUGUAGCGUUAUAUAAAUGCAAACUGUGUUGAUAUUUUUUAUGCUAAUUGCAUGCAAGUAAAACUUACACACACAU--ACA ((((((((.(((((....)))))....(((((((..........)))))))......)))))))).(((..(((((..(((((.....))))).))))).)))............--... ( -30.50) >consensus CAGUUUGCUGCUGCGGUGGCAGCUUCAAUGAUGCAUAAUUUGUAGCGUUAUAUAAAUGCAAACUGUGUUGAUAUUUUUUAUGCUAAUUGCAUGCAAGUAAAACUUACACACACAU_CACA ((((((((.(((((....)))))....(((((((..........)))))))......)))))))).(((..(((((..(((((.....))))).))))).)))................. (-29.08 = -29.48 + 0.40)

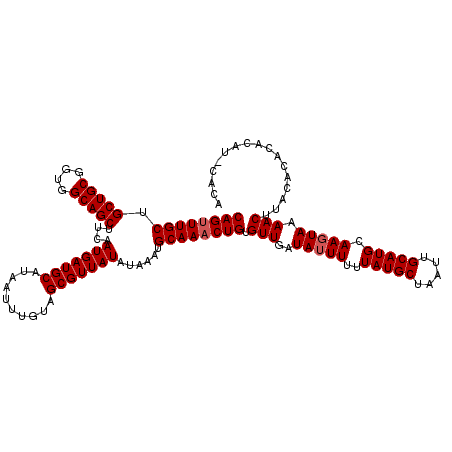
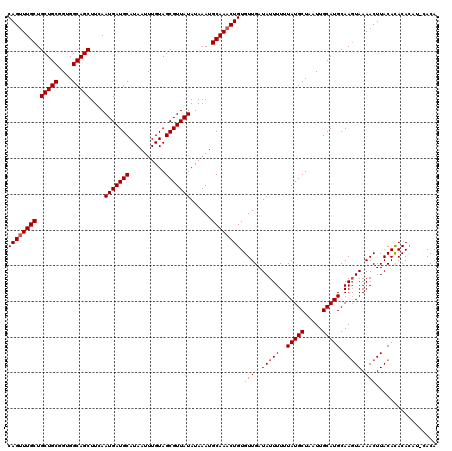
| Location | 10,540,432 – 10,540,548 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.49 |
| Mean single sequence MFE | -33.64 |
| Consensus MFE | -33.08 |
| Energy contribution | -32.96 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.90 |
| Structure conservation index | 0.98 |
| SVM decision value | 5.24 |
| SVM RNA-class probability | 0.999980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10540432 116 - 20766785 UGUG----UGUGUGUAAGUUUUACUUGCAUGCAAUUAGCAUAAAAAAUAUCAACACAGUUUGCAUUUAUAUAACGCUACAAAUUAUGCAUCAUUGAAGCUGCCACCGCAGCAGCAAACUG (((.----((((((((((.....))))))))))....)))...............((((((((...........((..........)).........(((((....))))).)))))))) ( -33.40) >DroSec_CAF1 33701 120 - 1 UGUGUAUGUGUGUGUAAGUUUUAGUUGCAUGCAAUUAGCAUAAAAAAUAUCAACACAGUUUGCAUUUAUAUAACGCUACAAAUUAUGCAUCAUUGAAGCUGCCACCGCAGCAGCAAACUG (((((...(((((((((.......)))))))))....))))).............((((((((...........((..........)).........(((((....))))).)))))))) ( -31.40) >DroSim_CAF1 30152 120 - 1 UGUGUAUGUGUGUGUAAGUUUUACUUGCAUGCAAUUAGCAUAAAAAAUAUCAACACAGUUUGCAUUUAUAUAACGCUACAAAUUAUGCAUCAUUGAAGCUGCCACCGCAGCAGCAAACUG (((((...((((((((((.....))))))))))....))))).............((((((((...........((..........)).........(((((....))))).)))))))) ( -35.50) >DroEre_CAF1 35100 118 - 1 UGU--GAGUGUAUGUAAGUUUUACUUGCAUGCAAUUAGCAUAAAAAAUAUCAACACAGUUUGCAUUUAUAUAACGCUACAAAUUAUGCAUCAUUGAAGCUGCCACCGCAGCAGCAAGCUG (((--...((((((((((.....))))))))))....)))...............((((((((...........((..........)).........(((((....))))).)))))))) ( -35.00) >DroYak_CAF1 34830 118 - 1 UGU--AUGUGUGUGUAAGUUUUACUUGCAUGCAAUUAGCAUAAAAAAUAUCAACACAGUUUGCAUUUAUAUAACGCUACAAAUUAUGCAUCAUUGAAGCUGCCACCGCAGCAGCAAACUG (((--...((((((((((.....))))))))))....)))...............((((((((...........((..........)).........(((((....))))).)))))))) ( -32.90) >consensus UGUG_AUGUGUGUGUAAGUUUUACUUGCAUGCAAUUAGCAUAAAAAAUAUCAACACAGUUUGCAUUUAUAUAACGCUACAAAUUAUGCAUCAUUGAAGCUGCCACCGCAGCAGCAAACUG (((.....((((((((((.....))))))))))....)))...............((((((((...........((..........)).........(((((....))))).)))))))) (-33.08 = -32.96 + -0.12)


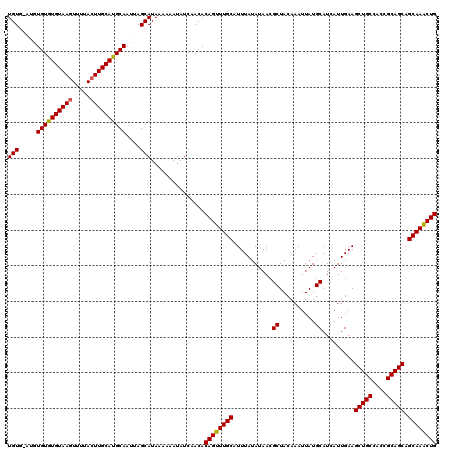
| Location | 10,540,548 – 10,540,650 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 90.87 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -26.74 |
| Energy contribution | -26.93 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10540548 102 + 20766785 UAGCCAGCAUAGCCUGUAAUUACAUGAACCCGUGGCGAAAAAAGAAAGUUGACAGUUAGGUGGCGUCAACGGCAGGUAGCAAACAAGUUGCAACUGCAACAG------ ......((...((((((.....((((....)))).............((((((.(((....))))))))).)))))).))......(((((....)))))..------ ( -27.90) >DroSec_CAF1 33821 100 + 1 UAGCCAGCU--CCCUGUAAUUACAUGAACCCGUGGCAAAAAAAGAAAGUUGACAGUUAGGUGGCGUCAACGGCAGGUAGCAAACAAGUUGCAACUGCAACAG------ ......(((--.(((((.....((((....)))).............((((((.(((....))))))))).))))).)))......(((((....)))))..------ ( -29.80) >DroSim_CAF1 30272 100 + 1 UAGCCAGCU--CCCUGUAAUUACAUGAACCCGUGGCGAAAAAAGAAAGUUGACAGUUAGGUGGCGUCAACGGCAGGUAGCAAACAAGUUGCAACUGCAACAG------ ......(((--.(((((.....((((....)))).............((((((.(((....))))))))).))))).)))......(((((....)))))..------ ( -29.80) >DroEre_CAF1 35218 104 + 1 CAGUCAGCU--CCCUGCAAUUACACGAGCCCG--GCGAAAAAAGAAAGUUGACAGUUAGGUGGCGUCAGCGGCAGGUAGCAAACAAGUUGCAACUGCAACAGCAGCGG ..((..(((--.(((((.......((....))--.............((((((.(((....))))))))).))))).)))..))..(((((....)))))........ ( -29.90) >consensus UAGCCAGCU__CCCUGUAAUUACAUGAACCCGUGGCGAAAAAAGAAAGUUGACAGUUAGGUGGCGUCAACGGCAGGUAGCAAACAAGUUGCAACUGCAACAG______ ......(((...(((((.....((((....)))).............((((((.(((....))))))))).))))).)))......(((((....)))))........ (-26.74 = -26.93 + 0.19)

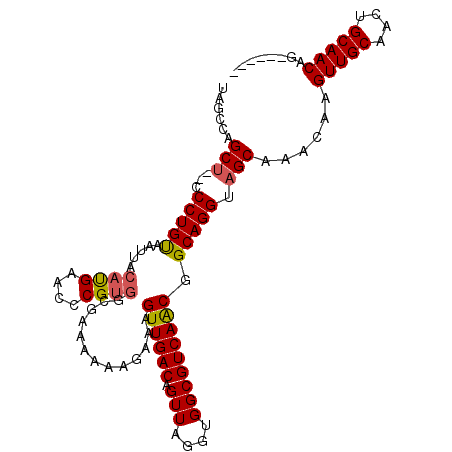
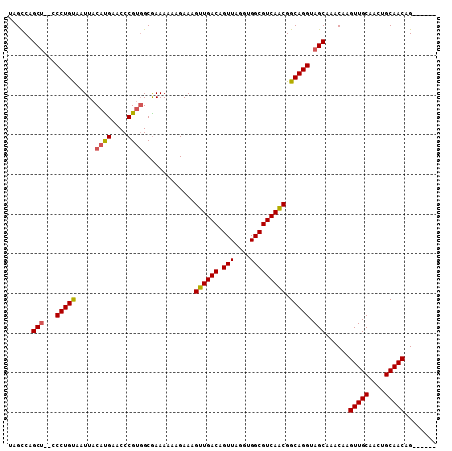
| Location | 10,540,588 – 10,540,687 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -28.61 |
| Energy contribution | -28.80 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10540588 99 + 20766785 AAAGAAAGUUGACAGUUAGGUGGCGUCAACGGCAGGUAGCAAACAAGUUGCAACUGCAACAG---------CAAUUGUUGUCGCUCCUUAUGACGCGACAGUAACGAG ..............((((.((.((((((...(((((((((......)))))..)))).((((---------(....))))).........)))))).))..))))... ( -29.10) >DroSec_CAF1 33859 99 + 1 AAAGAAAGUUGACAGUUAGGUGGCGUCAACGGCAGGUAGCAAACAAGUUGCAACUGCAACAG---------CAAUUGUUGUCGCUCCUUAUGACGCGACAGUAACGAG ..............((((.((.((((((...(((((((((......)))))..)))).((((---------(....))))).........)))))).))..))))... ( -29.10) >DroSim_CAF1 30310 99 + 1 AAAGAAAGUUGACAGUUAGGUGGCGUCAACGGCAGGUAGCAAACAAGUUGCAACUGCAACAG---------CAAUUGUUGUCGCUCCUUAUGACGCGACAGUAACGAG ..............((((.((.((((((...(((((((((......)))))..)))).((((---------(....))))).........)))))).))..))))... ( -29.10) >DroEre_CAF1 35254 108 + 1 AAAGAAAGUUGACAGUUAGGUGGCGUCAGCGGCAGGUAGCAAACAAGUUGCAACUGCAACAGCAGCGGCAGCAAUUGUUGUCGCUCCUUAUGACGCGACAGUAACGAG ..............((((.((.((((((((.(((((((((......)))))..))))....))(((((((((....))))))))).....)))))).))..))))... ( -41.30) >consensus AAAGAAAGUUGACAGUUAGGUGGCGUCAACGGCAGGUAGCAAACAAGUUGCAACUGCAACAG_________CAAUUGUUGUCGCUCCUUAUGACGCGACAGUAACGAG ..............((((.((.((((((.....(((.(((.((((((((((....))))).((.......))..)))))...))))))..)))))).))..))))... (-28.61 = -28.80 + 0.19)

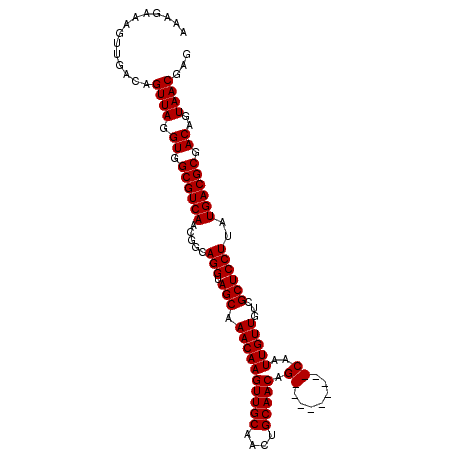
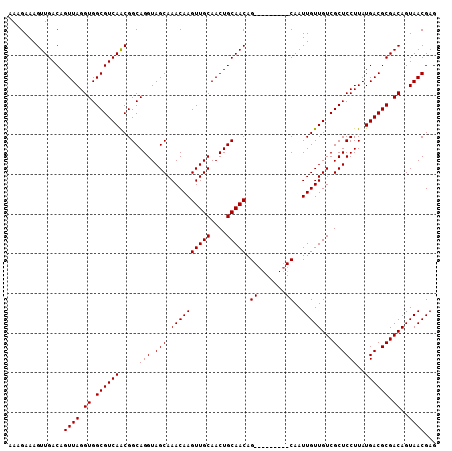
| Location | 10,540,588 – 10,540,687 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 95.17 |
| Mean single sequence MFE | -30.45 |
| Consensus MFE | -25.66 |
| Energy contribution | -25.65 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10540588 99 - 20766785 CUCGUUACUGUCGCGUCAUAAGGAGCGACAACAAUUG---------CUGUUGCAGUUGCAACUUGUUUGCUACCUGCCGUUGACGCCACCUAACUGUCAACUUUCUUU ...((((..((.((((((..(((((((((((((((((---------(....)))))))....))).))))).))).....)))))).)).)))).............. ( -29.60) >DroSec_CAF1 33859 99 - 1 CUCGUUACUGUCGCGUCAUAAGGAGCGACAACAAUUG---------CUGUUGCAGUUGCAACUUGUUUGCUACCUGCCGUUGACGCCACCUAACUGUCAACUUUCUUU ...((((..((.((((((..(((((((((((((((((---------(....)))))))....))).))))).))).....)))))).)).)))).............. ( -29.60) >DroSim_CAF1 30310 99 - 1 CUCGUUACUGUCGCGUCAUAAGGAGCGACAACAAUUG---------CUGUUGCAGUUGCAACUUGUUUGCUACCUGCCGUUGACGCCACCUAACUGUCAACUUUCUUU ...((((..((.((((((..(((((((((((((((((---------(....)))))))....))).))))).))).....)))))).)).)))).............. ( -29.60) >DroEre_CAF1 35254 108 - 1 CUCGUUACUGUCGCGUCAUAAGGAGCGACAACAAUUGCUGCCGCUGCUGUUGCAGUUGCAACUUGUUUGCUACCUGCCGCUGACGCCACCUAACUGUCAACUUUCUUU ...((((..((.((((((..((((((((..((((((((....(((((....))))).)))).))))))))).))).....)))))).)).)))).............. ( -33.00) >consensus CUCGUUACUGUCGCGUCAUAAGGAGCGACAACAAUUG_________CUGUUGCAGUUGCAACUUGUUUGCUACCUGCCGUUGACGCCACCUAACUGUCAACUUUCUUU ...((((..((.((((((..((((((((..((((..............(((((....)))))))))))))).))).....)))))).)).)))).............. (-25.66 = -25.65 + -0.00)

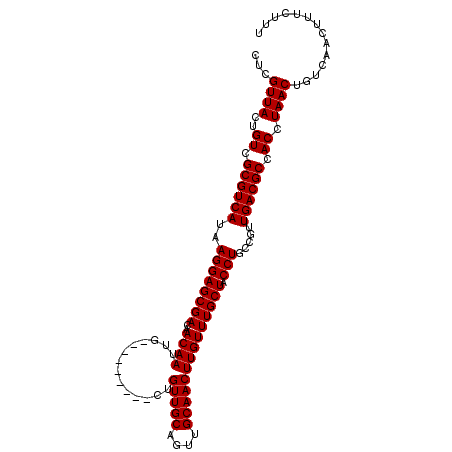
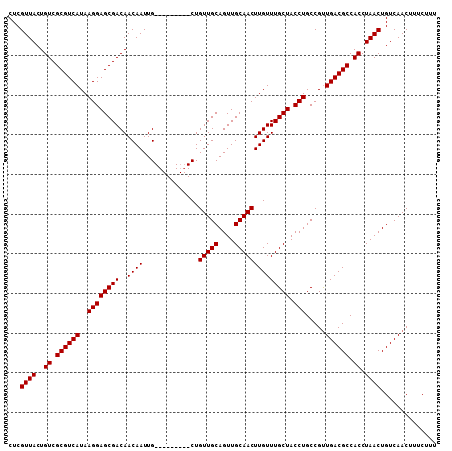
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:46 2006