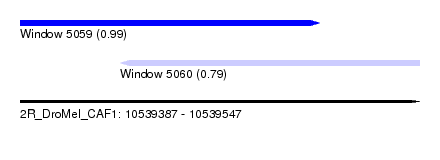
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,539,387 – 10,539,547 |
| Length | 160 |
| Max. P | 0.993296 |

| Location | 10,539,387 – 10,539,507 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.92 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -13.71 |
| Energy contribution | -14.63 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.45 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10539387 120 + 20766785 UCAUAGCUCUCCGGUGAUAUGUUCAAUACUUUCAAAUCGAAUGGUGUUACCACACCAUUGGCAAUUAUUUCAAGUUUUUCCAUUUGGCAGUGGUAAAAGCACUUCACCCGACUGCUGUGC .((((((..((.(((((..((((...((((.((((((.(((((((((....))))))((((........))))....))).)))))).)))).....))))..))))).))..)))))). ( -36.30) >DroSec_CAF1 32648 120 + 1 UCAUAGCUCUCCGGUGUUAUGUUUAGUACUUUCAAAUCGAAAGGAGUUACCACACCAUUGGCAAUUAUUUCAAGUUUUUCCAUUUGACAGUGGUAGAAGCACUUCACUCGACUGCUGUGC .((((((..((.((((...(((((....(((((.....)))))...(((((((.((...))........((((((......))))))..))))))))))))...)))).))..)))))). ( -31.70) >DroSim_CAF1 29075 120 + 1 UCAUAGCUCUCCGGUGAUAUGUUUAGUACUUUCAAAUCGAAAGGAGUUACCACACCAUUGGCAAUUAUUUCAAGUUUUUCCAUUUGGCAGUGAUAAAAGCACUUCACUCGACUGCUGUGC .((((((..((.(((((..(((((..((((.((((((.((((((((((.(((......))).))))........)))))).)))))).))))....)))))..))))).))..)))))). ( -31.70) >DroEre_CAF1 34453 95 + 1 UUGUAGUUUUCCUGUGUGUUAUUCGGAUUGGAUACAUCGAAUGGGUUGAUCACUCCAUUGUCAGCCACUUUUAGCUUUUCCAUCUGACAAUACA-----C-------------------- .............(((((((..((((((.(((......(((..((((((.((......))))))))...)))......))))))))).))))))-----)-------------------- ( -23.30) >DroYak_CAF1 32808 120 + 1 UCAUAGUUCUCCGGUGUUAUAUUGAGUACGUGCGGAUCAAAACUAGUCACCACACCAUUGGCAAUUAUUUCAAGUUUUUCCAUUUGGCAAUGGUAAAAACACUUCACCCGGCUGCUGUGU .((((((...((((.((......((((..(((..(((........)))..)))(((((((.(((...................))).)))))))......)))).))))))..)))))). ( -30.11) >consensus UCAUAGCUCUCCGGUGUUAUGUUCAGUACUUUCAAAUCGAAAGGAGUUACCACACCAUUGGCAAUUAUUUCAAGUUUUUCCAUUUGGCAGUGGUAAAAGCACUUCACCCGACUGCUGUGC .((((((..((.((((.(((.....))).........................(((((((.(((...................))).)))))))..........)))).))..)))))). (-13.71 = -14.63 + 0.92)

| Location | 10,539,427 – 10,539,547 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -24.26 |
| Energy contribution | -24.82 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10539427 120 - 20766785 AAAAACUGGGAAUUACCCCAGAUUACUUCGAAAAUUUUCCGCACAGCAGUCGGGUGAAGUGCUUUUACCACUGCCAAAUGGAAAAACUUGAAAUAAUUGCCAAUGGUGUGGUAACACCAU .....(((((......)))))((((.(((((...(((((((....(((((..(((((((...))))))))))))....)))))))..))))).)))).....(((((((....))))))) ( -37.40) >DroSec_CAF1 32688 120 - 1 AAAAACUGGGCAUUACCCCAGAUUACUUCGAAAAUUUUCCGCACAGCAGUCGAGUGAAGUGCUUCUACCACUGUCAAAUGGAAAAACUUGAAAUAAUUGCCAAUGGUGUGGUAACUCCUU .....(((((......)))))((((.(((((...(((((((....(((((.(.(((((....))).))))))))....)))))))..))))).))))(((((......)))))....... ( -28.10) >DroSim_CAF1 29115 120 - 1 AAAAACUGGGCAUUACCCCAGAUUACUUUGAAAAUUUUCCGCACAGCAGUCGAGUGAAGUGCUUUUAUCACUGCCAAAUGGAAAAACUUGAAAUAAUUGCCAAUGGUGUGGUAACUCCUU .....(((((......)))))((((.((..(...(((((((....(((((.((..(((....)))..)))))))....)))))))..)..)).))))(((((......)))))....... ( -26.30) >DroYak_CAF1 32848 120 - 1 AAAAACUGGGAAUUACCGAAGAUUACUUUGAGAACUUUCCACACAGCAGCCGGGUGAAGUGUUUUUACCAUUGCCAAAUGGAAAAACUUGAAAUAAUUGCCAAUGGUGUGGUGACUAGUU ...((((((..(((((....(((((.((..((...((((((....((((...(((((((...))))))).))))....))))))..))..)).)))))(((...))))))))..)))))) ( -30.60) >consensus AAAAACUGGGAAUUACCCCAGAUUACUUCGAAAAUUUUCCGCACAGCAGUCGAGUGAAGUGCUUUUACCACUGCCAAAUGGAAAAACUUGAAAUAAUUGCCAAUGGUGUGGUAACUCCUU .....(((((......))))).(((.(((((...(((((((....(((((..(((((((...))))))))))))....)))))))..))))).)))((((((......))))))...... (-24.26 = -24.82 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:38 2006