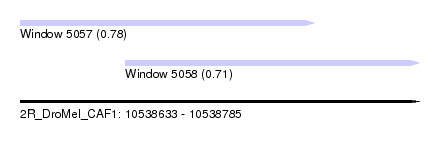
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,538,633 – 10,538,785 |
| Length | 152 |
| Max. P | 0.784379 |

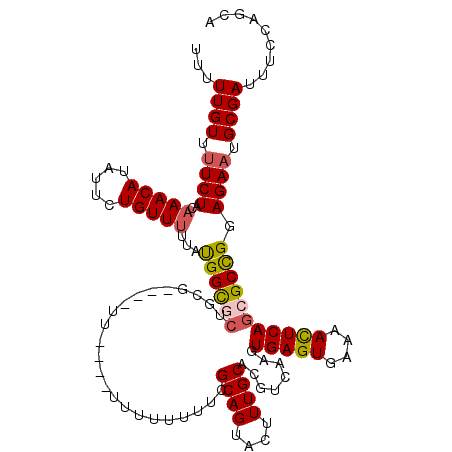
| Location | 10,538,633 – 10,538,745 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.18 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -21.70 |
| Energy contribution | -23.40 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10538633 112 + 20766785 UUUUUGUUUUCUACAAACAUAUUCUGUUUUUAUGGCGCUGCG----UU----UUUUUUUUCGCAGUACUUUGCACGUCAAGUGAGUGAAAAUUCAGCGCUGGAGAAUGCGAUUUCCAGCA ..(((((.....)))))((((.........))))((((((((----..----........))))))...............(((((....)))))))((((((((......)))))))). ( -29.30) >DroSec_CAF1 31871 112 + 1 UUUUUGUUUUCUACAAACAUAUUCUGUUUUUACGGCGCUGCG----UU----UUUUUUUUCGCAGUACUUUGCACGUCAAGUGAGUGAAAACUCAGCGCUGGAGAUUGCGAUUUCCAGCA ..(((((.....)))))........((((((((((..(((((----..----........)))))..))...(((.....))).)))))))).....(((((((((....))))))))). ( -33.70) >DroSim_CAF1 28266 111 + 1 UUUUUGUUUUCUACAAACAUAUUCUGUUUUUACGGCGCUGCG----UU----UUU-UUUUCGCAGUACUUUGCACGUCAAGUGAGUGAAAACUCAGCGCCGGAGAAUGCGAUUUCCAGCA .....((((((...(((((.....)))))...((((((((.(----((----(((-..(((((((....))))((.....))))).)))))).))))))))))))))((........)). ( -33.40) >DroEre_CAF1 33585 104 + 1 UUUUUGUUUUCUACGAACAUAUUCUGUU-UUAUGGC---------------GCUUUUUUUCGCAGUACUUUGCACGUGAAGUGAGUGAAAACUCAGAGCCGUAGAAUGCGAUUUCCAGCA ...((((.(((((((((((.....))))-)...(((---------------(((((.....((((....))))....)))))((((....))))...))))))))).))))......... ( -28.60) >DroYak_CAF1 32110 120 + 1 UUUUUGUUUUCUACAAACAUAUUCUGUUUUUAUGGUGCUGCUGCUGUUGCUGCUUUUUUUCGCAGUACUUUGCACGUCAAGUGAGUGAAAACUCAGAGCCGGAGAAUGCGAUUUCCAGCA ..(((((.....)))))...............((..(.(((....((.(((((........)))))))...))))..))..(((((....)))))..((.(((((......))))).)). ( -28.50) >consensus UUUUUGUUUUCUACAAACAUAUUCUGUUUUUAUGGCGCUGCG____UU____UUUUUUUUCGCAGUACUUUGCACGUCAAGUGAGUGAAAACUCAGCGCCGGAGAAUGCGAUUUCCAGCA ...((((.((((..(((((.....)))))...((((((.......................((((....))))........(((((....))))))))))).)))).))))......... (-21.70 = -23.40 + 1.70)


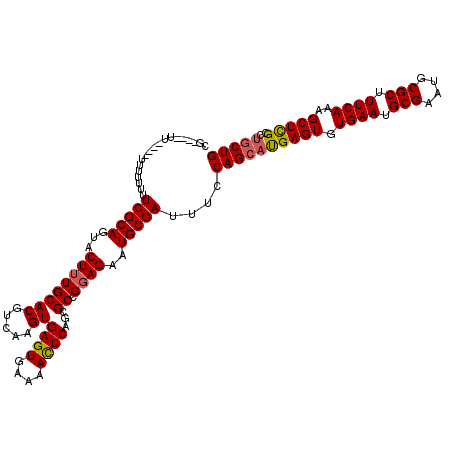
| Location | 10,538,673 – 10,538,785 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.78 |
| Mean single sequence MFE | -35.78 |
| Consensus MFE | -28.72 |
| Energy contribution | -30.34 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10538673 112 + 20766785 CG----UU----UUUUUUUUCGCAGUACUUUGCACGUCAAGUGAGUGAAAAUUCAGCGCUGGAGAAUGCGAUUUCCAGCAUGAGUGUGAAUGCGAAUGCGCUUUCAAAGCUUGGCUUCUG ..----..----.........((((....))))..(((((((...((((((((((..((((((((......)))))))).)))))......(((....))))))))..)))))))..... ( -35.70) >DroSec_CAF1 31911 112 + 1 CG----UU----UUUUUUUUCGCAGUACUUUGCACGUCAAGUGAGUGAAAACUCAGCGCUGGAGAUUGCGAUUUCCAGCAUGAGUGUGAAUGCGAAUGCGCUUUCAAAGCUCGCCUGCUG ..----..----.........((((...((..(((.(((..(((((....)))))..(((((((((....))))))))).))))))..)).((((....(((.....))))))))))).. ( -38.40) >DroSim_CAF1 28306 111 + 1 CG----UU----UUU-UUUUCGCAGUACUUUGCACGUCAAGUGAGUGAAAACUCAGCGCCGGAGAAUGCGAUUUCCAGCAUGAGUGUGAAUGCGAAUGCGCUUUCAAAGCUCGCCUGCUG ..----..----...-...(((((...(((((..(((....(((((....)))))))).)))))..)))))....((((((((((.((((.(((....))).))))..)))))..))))) ( -35.40) >DroEre_CAF1 33620 109 + 1 -----------GCUUUUUUUCGCAGUACUUUGCACGUGAAGUGAGUGAAAACUCAGAGCCGUAGAAUGCGAUUUCCAGCACGAGUGUGAAUGCGAAUGCGCUUUCAAAGCUCGCCAGCUG -----------........(((((....(((((..((....(((((....)))))..)).))))).)))))....((((.(((((.((((.(((....))).))))..)))))...)))) ( -32.70) >DroYak_CAF1 32150 120 + 1 CUGCUGUUGCUGCUUUUUUUCGCAGUACUUUGCACGUCAAGUGAGUGAAAACUCAGAGCCGGAGAAUGCGAUUUCCAGCAUGAGUGUGAAUGCGAAUGCGCUUUCAAAGCUCGUAUGCUG .(((...((((((........))))))....))).......(((((....)))))..((.(((((......))))).))((((((.((((.(((....))).))))..))))))...... ( -36.70) >consensus CG____UU____UUUUUUUUCGCAGUACUUUGCACGUCAAGUGAGUGAAAACUCAGCGCCGGAGAAUGCGAUUUCCAGCAUGAGUGUGAAUGCGAAUGCGCUUUCAAAGCUCGCCUGCUG ...................(((((...((((((((.....))((((....))))...)).))))..)))))....((((((((((.((((.(((....))).))))..)))))..))))) (-28.72 = -30.34 + 1.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:36 2006