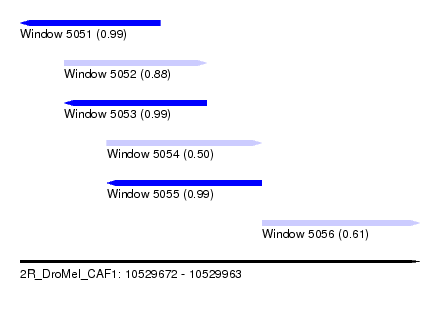
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,529,672 – 10,529,963 |
| Length | 291 |
| Max. P | 0.994602 |

| Location | 10,529,672 – 10,529,774 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.59 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -26.09 |
| Energy contribution | -26.01 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10529672 102 - 20766785 AACCGAUUAGCUUGGGCUUAAGCC-CAAGUAAGUUCAAUGCACUGCGAUUCAGUAUGCCGUUGAAGGAUUU-----------------AUGCUGAAAAGUUGUGUGCUAACACAUUUUAG ....((((.((((((((....)))-))))).))))....((((.((..(((((((((((......))...)-----------------))))))))..))...))))............. ( -33.10) >DroSec_CAF1 30308 103 - 1 AACCGAUUAGCUUGGGCUUAAGCCGCAAGUAAGUUCAAUGCACUACGAAUCAGUAUGCCGUUGAAGGAUUU-----------------AUGCUGAAAAGUUGUGUGCUAACACAUUUUAG ....((((.((((((((....))).))))).))))....((((.((((.((((((((((......))...)-----------------)))))))....))))))))............. ( -28.50) >DroSim_CAF1 26664 103 - 1 AACCGAUUAGCUUGGGCUUAAGCCCCAAGUAAGUUCAAUGCACUACGAAUCAGUAUGCCGUUGAAGGAUUU-----------------AUGCUGAAAAGUUGUGUGCUAACACAUUUUAG ....((((.((((((((....).))))))).))))....((((.((((.((((((((((......))...)-----------------)))))))....))))))))............. ( -28.50) >DroEre_CAF1 32018 119 - 1 AACCGAUUAGCUUGGGCUUAAGCC-CAAGUAAGUUCAAUGCACUGCAAUUCAGUAUGCCGGUGAAGGAUUUACUAUACCCGUGCUCGUAUGCUGAAAAGUUGUGUGCUAACACAUUUUAG ....((((.((((((((....)))-))))).))))....((((.((((((((((((((.((((..((((.....)).))..)))).)))))))))...)))))))))............. ( -39.80) >DroYak_CAF1 30520 102 - 1 AAGCGAUUAGCUUGGGCUUAAGCC-CAAGUAAGUUCAAUGCACUACAAUUCAGUAUGCCGGUGAAGGAUUU-----------------AUGCUGAAAAGUUGUGUGCUAACACAUUUUAG .(((((((.((((((((....)))-))))).))))....((((.((..(((((((((((......))...)-----------------))))))))..)).)))))))............ ( -34.10) >consensus AACCGAUUAGCUUGGGCUUAAGCC_CAAGUAAGUUCAAUGCACUACGAUUCAGUAUGCCGUUGAAGGAUUU_________________AUGCUGAAAAGUUGUGUGCUAACACAUUUUAG ....((((.((((((((....))).))))).))))....((((.((((((((((((.((......)).....................)))))))...)))))))))............. (-26.09 = -26.01 + -0.08)

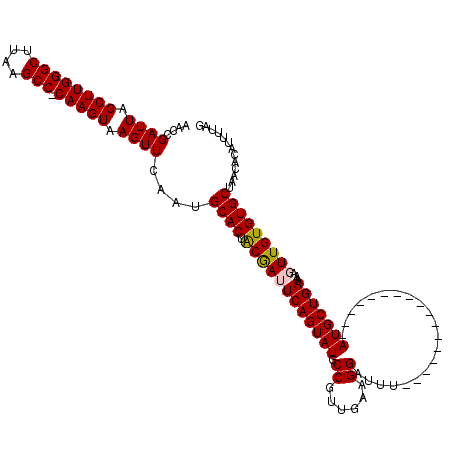
| Location | 10,529,704 – 10,529,808 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.88 |
| Mean single sequence MFE | -27.58 |
| Consensus MFE | -21.66 |
| Energy contribution | -21.86 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10529704 104 + 20766785 ---------AAAUCCUUCAACGGCAUACUGAAUCGCAGUGCAUUGAACUUACUUG-GGCUUAAGCCCAAGCUAAUCGGUUGCCCAACUACUGCCACUUGGCCAGCAGC------ACAUAC ---------............((((.(((((...((...))..........((((-(((....)))))))....)))))))))...((.(((((....)).))).)).------...... ( -28.10) >DroSec_CAF1 30340 102 + 1 ---------AAAUCCUUCAACGGCAUACUGAUUCGUAGUGCAUUGAACUUACUUGCGGCUUAAGCCCAAGCUAAUCGGUUGCCCAACUACUGCCACUUGGCCAG---C------ACAUAU ---------............((((.(((((((...(((.......)))..((((.(((....)))))))..)))))))))))......(((((....)).)))---.------...... ( -24.40) >DroSim_CAF1 26696 102 + 1 ---------AAAUCCUUCAACGGCAUACUGAUUCGUAGUGCAUUGAACUUACUUGGGGCUUAAGCCCAAGCUAAUCGGUUGCCCAACUACUGCCACUUGGCCAG---C------ACAUAU ---------............((((.(((((((.(((((.......)))......((((....))))..)).)))))))))))......(((((....)).)))---.------...... ( -26.00) >DroEre_CAF1 32058 109 + 1 GGGUAUAGUAAAUCCUUCACCGGCAUACUGAAUUGCAGUGCAUUGAACUUACUUG-GGCUUAAGCCCAAGCUAAUCGGUUGCCCAACUACUGCCACUCGGCCAG---C------ACAUA- (((((.............((((((..((((.....))))))..........((((-(((....))))))).....))))))))).....(((((....)).)))---.------.....- ( -30.21) >DroYak_CAF1 30552 106 + 1 ---------AAAUCCUUCACCGGCAUACUGAAUUGUAGUGCAUUGAACUUACUUG-GGCUUAAGCCCAAGCUAAUCGCUUGCCCAACUACUGCCACUCGGCCAG---CCAGCACACAUA- ---------............(((..........((((((((.(((..(((((((-(((....))))))).))))))..)))...))))).(((....)))..)---))..........- ( -29.20) >consensus _________AAAUCCUUCAACGGCAUACUGAAUCGUAGUGCAUUGAACUUACUUG_GGCUUAAGCCCAAGCUAAUCGGUUGCCCAACUACUGCCACUUGGCCAG___C______ACAUA_ .....................((((.(((((.....(((.......)))..((((.(((....)))))))....)))))))))......(((((....)).)))................ (-21.66 = -21.86 + 0.20)

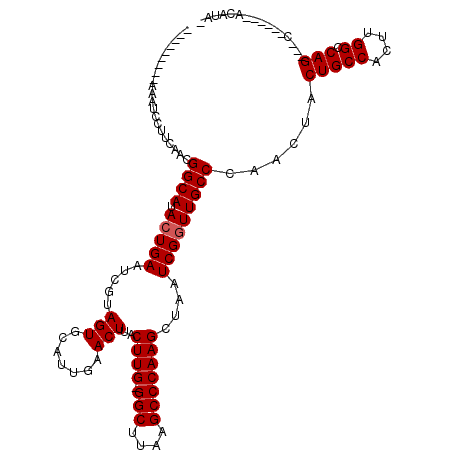
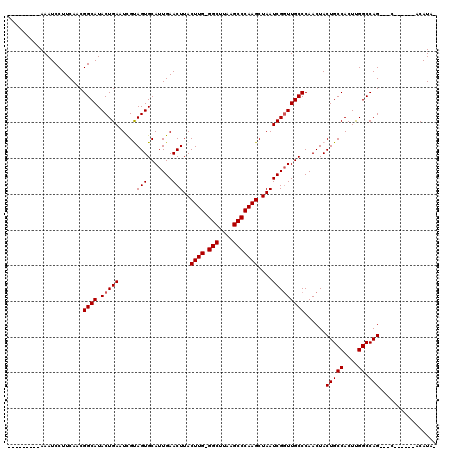
| Location | 10,529,704 – 10,529,808 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.88 |
| Mean single sequence MFE | -36.92 |
| Consensus MFE | -29.52 |
| Energy contribution | -29.88 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10529704 104 - 20766785 GUAUGU------GCUGCUGGCCAAGUGGCAGUAGUUGGGCAACCGAUUAGCUUGGGCUUAAGCC-CAAGUAAGUUCAAUGCACUGCGAUUCAGUAUGCCGUUGAAGGAUUU--------- .....(------((..((.....))..))).(((((((....)))))))((((((((....)))-)))))...(((((((((.(((......))))).)))))))......--------- ( -40.50) >DroSec_CAF1 30340 102 - 1 AUAUGU------G---CUGGCCAAGUGGCAGUAGUUGGGCAACCGAUUAGCUUGGGCUUAAGCCGCAAGUAAGUUCAAUGCACUACGAAUCAGUAUGCCGUUGAAGGAUUU--------- .....(------(---(((.((....)))))))(((((....)))))..((((((((....))).)))))...(((((((((.(((......))))).)))))))......--------- ( -33.20) >DroSim_CAF1 26696 102 - 1 AUAUGU------G---CUGGCCAAGUGGCAGUAGUUGGGCAACCGAUUAGCUUGGGCUUAAGCCCCAAGUAAGUUCAAUGCACUACGAAUCAGUAUGCCGUUGAAGGAUUU--------- .....(------(---(((.((....)))))))(((((....)))))..((((((((....).)))))))...(((((((((.(((......))))).)))))))......--------- ( -33.20) >DroEre_CAF1 32058 109 - 1 -UAUGU------G---CUGGCCGAGUGGCAGUAGUUGGGCAACCGAUUAGCUUGGGCUUAAGCC-CAAGUAAGUUCAAUGCACUGCAAUUCAGUAUGCCGGUGAAGGAUUUACUAUACCC -...(.------(---(..((.((((.(((((((((((....)))))).((((((((....)))-)))))...........))))).)))).))..)))((((.((......)).)))). ( -40.70) >DroYak_CAF1 30552 106 - 1 -UAUGUGUGCUGG---CUGGCCGAGUGGCAGUAGUUGGGCAAGCGAUUAGCUUGGGCUUAAGCC-CAAGUAAGUUCAAUGCACUACAAUUCAGUAUGCCGGUGAAGGAUUU--------- -.......(((((---(..(((....))).(((((...(((...((((.((((((((....)))-))))).))))...))))))))..........)))))).........--------- ( -37.00) >consensus _UAUGU______G___CUGGCCAAGUGGCAGUAGUUGGGCAACCGAUUAGCUUGGGCUUAAGCC_CAAGUAAGUUCAAUGCACUACGAUUCAGUAUGCCGUUGAAGGAUUU_________ ...................(((....)))..(((((((....)))))))((((((((....))).)))))...(((((((((.(((......))))).)))))))............... (-29.52 = -29.88 + 0.36)

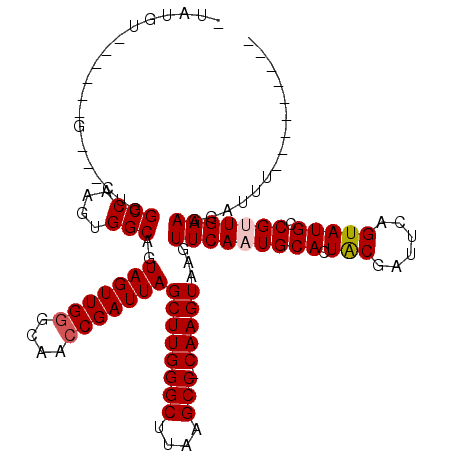
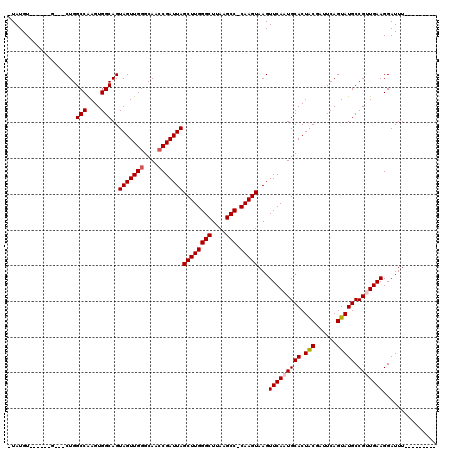
| Location | 10,529,735 – 10,529,848 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.52 |
| Mean single sequence MFE | -27.26 |
| Consensus MFE | -16.46 |
| Energy contribution | -16.86 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10529735 113 + 20766785 CAUUGAACUUACUUG-GGCUUAAGCCCAAGCUAAUCGGUUGCCCAACUACUGCCACUUGGCCAGCAGC------ACAUACAGCACAUAUGUAGUGUGUUAAAGCCACUUGCUCUGAUUAA ...........((((-(((....))))))).(((((((..((.......(((((....)).))).(((------(((((((.......))).)))))))..........)).))))))). ( -32.40) >DroSec_CAF1 30371 110 + 1 CAUUGAACUUACUUGCGGCUUAAGCCCAAGCUAAUCGGUUGCCCAACUACUGCCACUUGGCCAG---C------ACAUAU-GUAUGUAGUUAGUAUGUUAAAGCCACUUGCUCUGAUUAA ...........((((.(((....))))))).(((((((.(((..((((((.(((....)))..(---(------(....)-))..)))))).)))......(((.....)))))))))). ( -24.50) >DroSim_CAF1 26727 110 + 1 CAUUGAACUUACUUGGGGCUUAAGCCCAAGCUAAUCGGUUGCCCAACUACUGCCACUUGGCCAG---C------ACAUAU-GUAUGUAGUUAGUAUGUUAAAGCCACUUGCUCUGAUUAA ...........((((((.(....))))))).(((((((.(((..((((((.(((....)))..(---(------(....)-))..)))))).)))......(((.....)))))))))). ( -24.50) >DroEre_CAF1 32098 90 + 1 CAUUGAACUUACUUG-GGCUUAAGCCCAAGCUAAUCGGUUGCCCAACUACUGCCACUCGGCCAG---C------ACAUA-------G-------------UAGCCACUUGCUCUGAUUAA ...........((((-(((....))))))).(((((((..((....((((((((....)))...---.------....)-------)-------------)))......)).))))))). ( -25.50) >DroYak_CAF1 30583 99 + 1 CAUUGAACUUACUUG-GGCUUAAGCCCAAGCUAAUCGCUUGCCCAACUACUGCCACUCGGCCAG---CCAGCACACAUA-------G----AGU------GAGGCACUUGCUCUGAUUAA ...........((((-(((....))))))).((((((((.((.........(((....)))..)---).)))......(-------(----((.------.((...))..))))))))). ( -29.40) >consensus CAUUGAACUUACUUG_GGCUUAAGCCCAAGCUAAUCGGUUGCCCAACUACUGCCACUUGGCCAG___C______ACAUA__G_A__UA__UAGU_UGUUAAAGCCACUUGCUCUGAUUAA ...........((((.(((....))))))).(((((((...........(((((....)).))).....................................(((.....)))))))))). (-16.46 = -16.86 + 0.40)

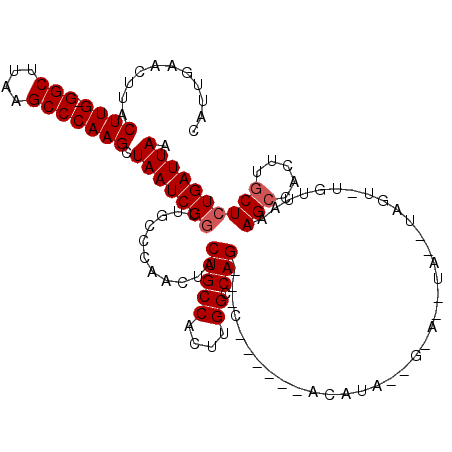
| Location | 10,529,735 – 10,529,848 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.52 |
| Mean single sequence MFE | -36.13 |
| Consensus MFE | -26.28 |
| Energy contribution | -26.48 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10529735 113 - 20766785 UUAAUCAGAGCAAGUGGCUUUAACACACUACAUAUGUGCUGUAUGU------GCUGCUGGCCAAGUGGCAGUAGUUGGGCAACCGAUUAGCUUGGGCUUAAGCC-CAAGUAAGUUCAAUG .......((((.((..((....((((((.......))).)))....------))..)).(((....)))..(((((((....)))))))((((((((....)))-)))))..)))).... ( -43.00) >DroSec_CAF1 30371 110 - 1 UUAAUCAGAGCAAGUGGCUUUAACAUACUAACUACAUAC-AUAUGU------G---CUGGCCAAGUGGCAGUAGUUGGGCAACCGAUUAGCUUGGGCUUAAGCCGCAAGUAAGUUCAAUG .......((((...(((((...(((((............-.)))))------.---..))))).(((((..(((((((....)))))))((....))....)))))......)))).... ( -34.42) >DroSim_CAF1 26727 110 - 1 UUAAUCAGAGCAAGUGGCUUUAACAUACUAACUACAUAC-AUAUGU------G---CUGGCCAAGUGGCAGUAGUUGGGCAACCGAUUAGCUUGGGCUUAAGCCCCAAGUAAGUUCAAUG .......((((...(((((...(((((............-.)))))------.---..)))))........(((((((....)))))))((((((((....).)))))))..)))).... ( -33.62) >DroEre_CAF1 32098 90 - 1 UUAAUCAGAGCAAGUGGCUA-------------C-------UAUGU------G---CUGGCCGAGUGGCAGUAGUUGGGCAACCGAUUAGCUUGGGCUUAAGCC-CAAGUAAGUUCAAUG .......((((.....((((-------------(-------(.((.------(---....)))))))))..(((((((....)))))))((((((((....)))-)))))..)))).... ( -35.30) >DroYak_CAF1 30583 99 - 1 UUAAUCAGAGCAAGUGCCUC------ACU----C-------UAUGUGUGCUGG---CUGGCCGAGUGGCAGUAGUUGGGCAAGCGAUUAGCUUGGGCUUAAGCC-CAAGUAAGUUCAAUG .......((((...(((((.------(((----.-------..(((.((((((---....)).)))))))..))).)))))........((((((((....)))-)))))..)))).... ( -34.30) >consensus UUAAUCAGAGCAAGUGGCUUUAACA_ACUA__UA__U_C__UAUGU______G___CUGGCCAAGUGGCAGUAGUUGGGCAACCGAUUAGCUUGGGCUUAAGCC_CAAGUAAGUUCAAUG .......((((................................................(((....)))..(((((((....)))))))((((((((....))).)))))..)))).... (-26.28 = -26.48 + 0.20)

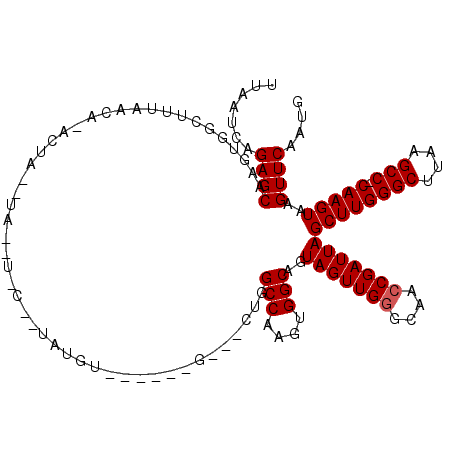
| Location | 10,529,848 – 10,529,963 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.15 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -20.27 |
| Energy contribution | -20.11 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10529848 115 + 20766785 AAGCGGCAAAAAAGCACGCAGAAAGGAGUUAAAAAAAAUGAGUGCAAAUUAUAUGACGAACGGGUUAUGGGU-----UGCGUACGAGCACUCUAAGCCAUAAAGAGCAAAUGAUGUUGGU ...((((......((((.((..................)).))))..(((((.((.......((((.(((..-----(((......)))..)))))))........)).))))))))).. ( -21.73) >DroSec_CAF1 30481 114 + 1 AAGCGGCAAAAAAGCACGCAGAAAGGAGUUAAA-AAAAUGAGUGCAAAUUAUAUGACGAACGGGUUAUGGGU-----UGCGUACCAGCACUCUAAGCCAUAAAGAGCAAAUGAUGUUGGU ..(((((......((((.((.............-....)).))))......((((((......)))))).))-----)))..(((((((.((...((........))....))))))))) ( -25.03) >DroSim_CAF1 26837 114 + 1 AAGCGGCAAAAAAGCACGCAGAAAGGAGUUAAA-AAAAUGAGUGCAAAUUAUAUGACGAACGGGUUAUGGGU-----UGCGUACAAGCACUCUAAGCCAUAAAGAGCAAAUGAUGUUGGU ...((((......((((.((.............-....)).))))..(((((.((.......((((.(((..-----(((......)))..)))))))........)).))))))))).. ( -21.79) >DroEre_CAF1 32188 110 + 1 AAGCGGCAAA-AAGCACGCAGAA--GAGUUAAA--AAAUGAGUGCAAAUUAUAUGACGAACGGGUUAUGGGU-----UGCGUACGAGCACUGUGAGCCGUAAAGAGCAUAUGAUGUUGGU ...((((...-..((((.((...--........--...)).))))..((((((((....((((.((((((..-----(((......))))))))).))))......)))))))))))).. ( -28.84) >DroYak_CAF1 30682 118 + 1 AAGCGGCAAAAAAGCACGCAGAAAGGAGUUAAA--AAAUGAGUGCAAAUUAUAUGACGAACGGGUUAUGGGUUGGCUUGCGUACGAGCACUAUAAGCCAUAAAGAGCAAAUGAUGUUGGU ...(((((.....((((.((.............--...)).))))................((.((((((....(((((....))))).)))))).))...............))))).. ( -25.19) >consensus AAGCGGCAAAAAAGCACGCAGAAAGGAGUUAAA_AAAAUGAGUGCAAAUUAUAUGACGAACGGGUUAUGGGU_____UGCGUACGAGCACUCUAAGCCAUAAAGAGCAAAUGAUGUUGGU ...((((......((((.((..................)).))))..(((((.((.(...(...((((((.(.....(((......))).....).)))))).).))).))))))))).. (-20.27 = -20.11 + -0.16)

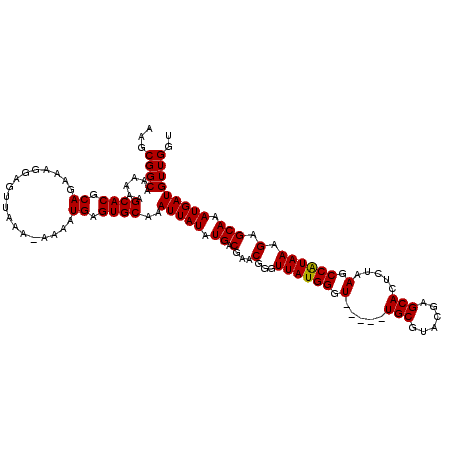
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:34 2006