| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,524,787 – 10,524,941 |
| Length | 154 |
| Max. P | 0.989515 |

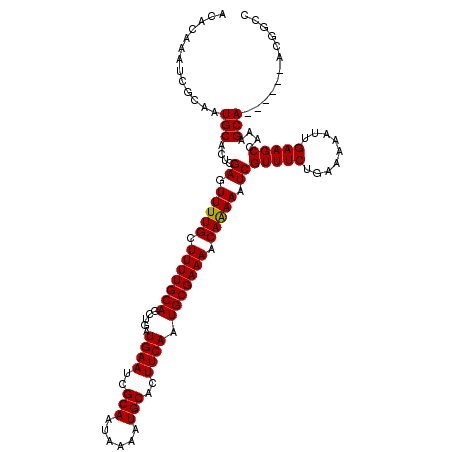
| Location | 10,524,787 – 10,524,901 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.05 |
| Mean single sequence MFE | -23.92 |
| Consensus MFE | -21.84 |
| Energy contribution | -21.68 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10524787 114 - 20766785 ACACAAAUCGCAAUGCACUCGAGUUUUGCUUUUGCAGUUGAUGAAUCGCAAUAAAAUGCACUUCAAUGCGAAAACAAAAAUCGUUUCUGAAAAAUUGAAGCCAAAGCA------ACGGCC .........((..(((....((.(((((.(((((((.....((((..(((......)))..)))).))))))).))))).))(((((.........)))))....)))------...)). ( -24.30) >DroSec_CAF1 25671 114 - 1 ACACAAAUCGCAAUGCACUCGAGUUUUGCUUUUGCAGCUGAUGAAUCGCAAUAAAAUGCACUUCAAUGCGAAAACAAAAAUCGUUUCUGAAAAAUUGAAGCCAAAGCA------ACGACC .......(((...(((....((.(((((.(((((((.....((((..(((......)))..)))).))))))).))))).))(((((.........)))))....)))------.))).. ( -23.40) >DroSim_CAF1 21855 114 - 1 ACACAAAUCGCAAUGCACUCGAGUUUUGCUUUUGCAGCUGAUGAAUCGCAAUAAAAUGCACUUCAAUGCGAAAACAAAAAUCGUUUCUGAAAAAUUGAAGCCAAAGCA------ACGGCC .........((..(((....((.(((((.(((((((.....((((..(((......)))..)))).))))))).))))).))(((((.........)))))....)))------...)). ( -24.30) >DroEre_CAF1 27914 114 - 1 ACACAAAUCGCAAUGCACUCGAGUUUUGCUUUUGCAGCUGAUGAAUCGCAAUAAAAUGCACUUCAAUGCGAAAACAAAAAUCGUUUCUGAAAAAUUGAAGCCAAAGCA------AUAGCC .........((..(((....((.(((((.(((((((.....((((..(((......)))..)))).))))))).))))).))(((((.........)))))....)))------...)). ( -23.20) >DroYak_CAF1 25971 120 - 1 ACACAAAUCGCAAUGCACUCGAGUUUUGCUUUUGCAGCUGAUGAAUCGCAAUAAAAUGCACUUCAAUGCGAAAACAGAAAUCGUUUCUGAAAAAUUGAAGCCAAAGCAACAGCAACAGCC .........((..((((...(((.....))).))))((((.((....(((......))).(((((((.......(((((.....)))))....)))))))......)).))))....)). ( -24.40) >consensus ACACAAAUCGCAAUGCACUCGAGUUUUGCUUUUGCAGCUGAUGAAUCGCAAUAAAAUGCACUUCAAUGCGAAAACAAAAAUCGUUUCUGAAAAAUUGAAGCCAAAGCA______ACGGCC .............(((....((.(((((.(((((((.....((((..(((......)))..)))).))))))).))))).))(((((.........)))))....)))............ (-21.84 = -21.68 + -0.16)


| Location | 10,524,821 – 10,524,941 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.33 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -27.52 |
| Energy contribution | -27.72 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10524821 120 + 20766785 UUUUUGUUUUCGCAUUGAAGUGCAUUUUAUUGCGAUUCAUCAACUGCAAAAGCAAAACUCGAGUGCAUUGCGAUUUGUGUUGUUCGAGAUGUGCGAGGUAUUUAAUUAAUUUGAAAAUAA .(((((((((.(((.((((.((((......)))).)))).....))).)))))))))((((...((((..(((..........)))..)))).))))...(((((.....)))))..... ( -27.90) >DroSec_CAF1 25705 120 + 1 UUUUUGUUUUCGCAUUGAAGUGCAUUUUAUUGCGAUUCAUCAGCUGCAAAAGCAAAACUCGAGUGCAUUGCGAUUUGUGUUGUUCGAGAUGUGCGAGGUAUUUAAUUAAUUUGAAAAUAA ...((((((((((..((((.((((......)))).))))...))(((....(((...(((((..((((........))))...)))))...)))...)))............)))))))) ( -28.70) >DroSim_CAF1 21889 120 + 1 UUUUUGUUUUCGCAUUGAAGUGCAUUUUAUUGCGAUUCAUCAGCUGCAAAAGCAAAACUCGAGUGCAUUGCGAUUUGUGUUGUUCGAGAUGUGCGAGGUAUUUAAUUAAUUUGAAAAUAA ...((((((((((..((((.((((......)))).))))...))(((....(((...(((((..((((........))))...)))))...)))...)))............)))))))) ( -28.70) >DroEre_CAF1 27948 120 + 1 UUUUUGUUUUCGCAUUGAAGUGCAUUUUAUUGCGAUUCAUCAGCUGCAAAAGCAAAACUCGAGUGCAUUGCGAUUUGUGUUGUUCGAGAUGUGCGAGGUAUUUAAUUAAUUUGAAAAUAA ...((((((((((..((((.((((......)))).))))...))(((....(((...(((((..((((........))))...)))))...)))...)))............)))))))) ( -28.70) >DroYak_CAF1 26011 120 + 1 UUUCUGUUUUCGCAUUGAAGUGCAUUUUAUUGCGAUUCAUCAGCUGCAAAAGCAAAACUCGAGUGCAUUGCGAUUUGUGUUGUUCGAGAUGUGCGAGGUAUUUAAUUAAUUUGAAAAUAA ....(((((((((..((((.((((......)))).))))...))(((....(((...(((((..((((........))))...)))))...)))...)))............))))))). ( -28.50) >consensus UUUUUGUUUUCGCAUUGAAGUGCAUUUUAUUGCGAUUCAUCAGCUGCAAAAGCAAAACUCGAGUGCAUUGCGAUUUGUGUUGUUCGAGAUGUGCGAGGUAUUUAAUUAAUUUGAAAAUAA ....(((((((((..((((.((((......)))).))))...))(((....(((...(((((..((((........))))...)))))...)))...)))............))))))). (-27.52 = -27.72 + 0.20)



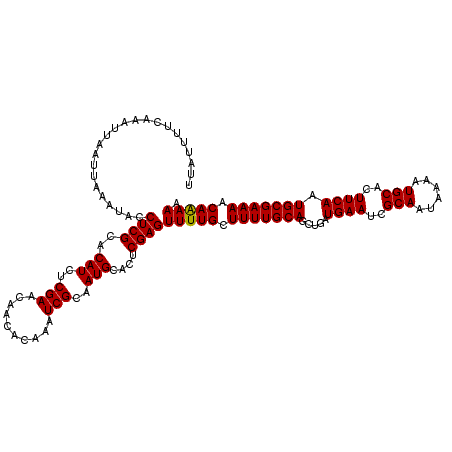
| Location | 10,524,821 – 10,524,941 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.33 |
| Mean single sequence MFE | -19.00 |
| Consensus MFE | -18.88 |
| Energy contribution | -18.72 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10524821 120 - 20766785 UUAUUUUCAAAUUAAUUAAAUACCUCGCACAUCUCGAACAACACAAAUCGCAAUGCACUCGAGUUUUGCUUUUGCAGUUGAUGAAUCGCAAUAAAAUGCACUUCAAUGCGAAAACAAAAA ........................(((((.........((((.((((..((((.((......)).)))).))))..)))).((((..(((......)))..)))).)))))......... ( -20.10) >DroSec_CAF1 25705 120 - 1 UUAUUUUCAAAUUAAUUAAAUACCUCGCACAUCUCGAACAACACAAAUCGCAAUGCACUCGAGUUUUGCUUUUGCAGCUGAUGAAUCGCAAUAAAAUGCACUUCAAUGCGAAAACAAAAA .......................((((..(((..(((..........)))..)))....))))(((((.(((((((.....((((..(((......)))..)))).))))))).))))). ( -18.70) >DroSim_CAF1 21889 120 - 1 UUAUUUUCAAAUUAAUUAAAUACCUCGCACAUCUCGAACAACACAAAUCGCAAUGCACUCGAGUUUUGCUUUUGCAGCUGAUGAAUCGCAAUAAAAUGCACUUCAAUGCGAAAACAAAAA .......................((((..(((..(((..........)))..)))....))))(((((.(((((((.....((((..(((......)))..)))).))))))).))))). ( -18.70) >DroEre_CAF1 27948 120 - 1 UUAUUUUCAAAUUAAUUAAAUACCUCGCACAUCUCGAACAACACAAAUCGCAAUGCACUCGAGUUUUGCUUUUGCAGCUGAUGAAUCGCAAUAAAAUGCACUUCAAUGCGAAAACAAAAA .......................((((..(((..(((..........)))..)))....))))(((((.(((((((.....((((..(((......)))..)))).))))))).))))). ( -18.70) >DroYak_CAF1 26011 120 - 1 UUAUUUUCAAAUUAAUUAAAUACCUCGCACAUCUCGAACAACACAAAUCGCAAUGCACUCGAGUUUUGCUUUUGCAGCUGAUGAAUCGCAAUAAAAUGCACUUCAAUGCGAAAACAGAAA .......................((((..(((..(((..........)))..)))....))))(((((.(((((((.....((((..(((......)))..)))).))))))).))))). ( -18.80) >consensus UUAUUUUCAAAUUAAUUAAAUACCUCGCACAUCUCGAACAACACAAAUCGCAAUGCACUCGAGUUUUGCUUUUGCAGCUGAUGAAUCGCAAUAAAAUGCACUUCAAUGCGAAAACAAAAA .......................((((..(((..(((..........)))..)))....))))(((((.(((((((.....((((..(((......)))..)))).))))))).))))). (-18.88 = -18.72 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:18 2006