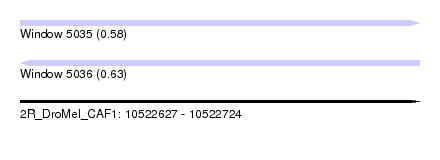
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,522,627 – 10,522,724 |
| Length | 97 |
| Max. P | 0.628386 |

| Location | 10,522,627 – 10,522,724 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -22.76 |
| Consensus MFE | -19.96 |
| Energy contribution | -20.24 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10522627 97 + 20766785 UUGCCAUGCCUGUGUAAAUAAUGCCUUUAUUAUUUCGCUUGAUAUCCAC-UCGCAGCGGAUAAAAAUGGCAGGGCAACGAAACCACAACAAGCAGCAG (((((.((((.(((.((((((((....)))))))))))....(((((.(-.....).))))).....)))).)))))..................... ( -24.70) >DroSec_CAF1 23535 97 + 1 UUGCCAUGCCUGUGUAAAUAAUGCCUUUAUUAUUUCGCUUGAUAUCCAC-UCGCACCGGAUAAAAAUGGCAGGGCAACGAAACCACAACAAGCAGCAG (((((.((((.(((.((((((((....)))))))))))....(((((..-.......))))).....)))).)))))..................... ( -23.30) >DroSim_CAF1 19654 98 + 1 UUGCCAUGCCUGUGUAAAUAAUGCCUUUAUUAUUUCGCUUGAUAUCCACCUCGCAGCGGAUAAAAAUGGCAGGGCAACGAAACCACAACAAGCAGCAG (((((.((((.(((.((((((((....)))))))))))....(((((.(......).))))).....)))).)))))..................... ( -24.20) >DroEre_CAF1 25838 97 + 1 UUGCCUCGCCUGUGUAAAUAAUGCCUUUAUUAUUUCGCUUGAUAUCCAC-UAACAGCAGAUAAAAAUGGCAGGGCAACGCAACCACAACAAGCAGCAG (((((..(((.(((.((((((((....)))))))))))....((((..(-.....)..)))).....)))..))))).((...........))..... ( -20.40) >DroYak_CAF1 23752 97 + 1 UUGCCAUGCCUGUGUAAAUAAUGCCUUUAUUAUUUUGCUUGAUAUCCAC-UCGCAGCAGAUAAAAAUGGCAGGGCAGCGAAACCACAACAAGCAGCAG .(((..(((.(((((......((((.((.((((((.((((((.......-))).))))))))).)).))))((.........)))).))).)))))). ( -21.20) >consensus UUGCCAUGCCUGUGUAAAUAAUGCCUUUAUUAUUUCGCUUGAUAUCCAC_UCGCAGCGGAUAAAAAUGGCAGGGCAACGAAACCACAACAAGCAGCAG (((((.((((.(((.((((((((....)))))))))))....(((((..........))))).....)))).)))))..................... (-19.96 = -20.24 + 0.28)

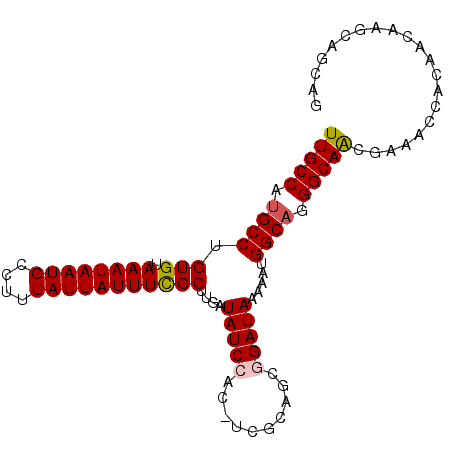
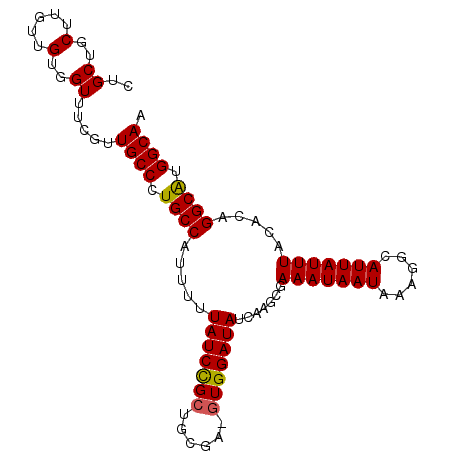
| Location | 10,522,627 – 10,522,724 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -26.34 |
| Consensus MFE | -25.26 |
| Energy contribution | -25.06 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.628386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10522627 97 - 20766785 CUGCUGCUUGUUGUGGUUUCGUUGCCCUGCCAUUUUUAUCCGCUGCGA-GUGGAUAUCAAGCGAAAUAAUAAAGGCAUUAUUUACACAGGCAUGGCAA ..((..(.....)..))....(((((.((((.....(((((((.....-)))))))....(.((((((((......))))))).).).)))).))))) ( -27.90) >DroSec_CAF1 23535 97 - 1 CUGCUGCUUGUUGUGGUUUCGUUGCCCUGCCAUUUUUAUCCGGUGCGA-GUGGAUAUCAAGCGAAAUAAUAAAGGCAUUAUUUACACAGGCAUGGCAA .((((((((((.(((((((((((......((((((.(((...))).))-))))......))))))))(((((.....))))))))))))))..)))). ( -24.90) >DroSim_CAF1 19654 98 - 1 CUGCUGCUUGUUGUGGUUUCGUUGCCCUGCCAUUUUUAUCCGCUGCGAGGUGGAUAUCAAGCGAAAUAAUAAAGGCAUUAUUUACACAGGCAUGGCAA ..((..(.....)..))....(((((.((((.....((((((((....))))))))....(.((((((((......))))))).).).)))).))))) ( -29.10) >DroEre_CAF1 25838 97 - 1 CUGCUGCUUGUUGUGGUUGCGUUGCCCUGCCAUUUUUAUCUGCUGUUA-GUGGAUAUCAAGCGAAAUAAUAAAGGCAUUAUUUACACAGGCGAGGCAA ..((.(((......))).)).(((((.((((.....((((..(.....-)..))))....(.((((((((......))))))).).).)))).))))) ( -25.30) >DroYak_CAF1 23752 97 - 1 CUGCUGCUUGUUGUGGUUUCGCUGCCCUGCCAUUUUUAUCUGCUGCGA-GUGGAUAUCAAGCAAAAUAAUAAAGGCAUUAUUUACACAGGCAUGGCAA ..((..(.....)..)).....((((.((((.....((((..(.....-)..)))).......(((((((......))))))).....)))).)))). ( -24.50) >consensus CUGCUGCUUGUUGUGGUUUCGUUGCCCUGCCAUUUUUAUCCGCUGCGA_GUGGAUAUCAAGCGAAAUAAUAAAGGCAUUAUUUACACAGGCAUGGCAA ..((..(.....)..)).....((((.((((.....(((((((......))))))).......(((((((......))))))).....)))).)))). (-25.26 = -25.06 + -0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:15 2006