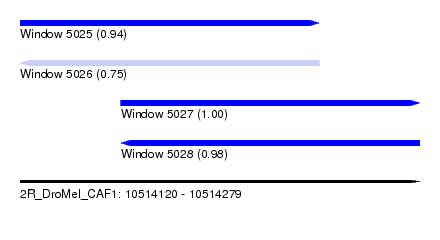
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,514,120 – 10,514,279 |
| Length | 159 |
| Max. P | 0.999887 |

| Location | 10,514,120 – 10,514,239 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.99 |
| Mean single sequence MFE | -22.52 |
| Consensus MFE | -19.98 |
| Energy contribution | -20.78 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10514120 119 + 20766785 CAAACAUUUUACGGUUUCACCUCGAAAUUAAAUUUCCCCU-UUCAAUACCGACCAAAAAAGCCGCAUCGAUAGACGGCCAAAACGUGGCAUCAUGGUGAAAACUAAAAUCAAAUUGCCAC ...........((((........(((((...)))))....-......)))).........((((..((....))))))......((((((...((((..........))))...)))))) ( -20.87) >DroSec_CAF1 14706 120 + 1 CAAACAUUUUACGGUUUCACCUCGAAAUUAAAUUUCCCCUUUUCAAUACCGGCCAAAAAAGCCGCAUCGAUAGACGGCCAAAACGUGGCAUCAUGGUGAAAACUAAAAUCAAAUUGCUAC .....((((((...(((((((..((((.............)))).......((((.....((((..((....)))))).......)))).....)))))))..))))))........... ( -23.32) >DroSim_CAF1 9727 120 + 1 CAAACAUUUUACGGUUUCACCUCGAAAUUAAAUUUCCCCUUUUCAAUACCGGCCAAAAAAGCCGCAUCGAUAGACGGCCAAAACGUGGCAUCAUGGUGAAAACUAAAAUCAAAUUGCCAC .....((((((...(((((((..((((.............)))).......((((.....((((..((....)))))).......)))).....)))))))..))))))........... ( -23.32) >DroEre_CAF1 14852 119 + 1 CAAACACUUUACGGUCUCACCUCGAAAUUAAAUUUCCCCU-UUCAAUGCCCGCCAAAAAAGCCGCAUCGAUAGACGGCCAAAACGUGGCAUCAUGGUGAAAACUAAAAUCAAAUUGCCAC ............((((.......(((((...)))))..((-.((.((((..((.......)).)))).)).))..)))).....((((((...((((..........))))...)))))) ( -22.90) >DroYak_CAF1 11418 118 + 1 CAAACACUUUACGGUUUCACCUCGAAAUUAAAUUUCCCCU-UUCAAUAUCGGCCAA-AAAGCCGCAUCGAUAGACGGCCCGAACGUGGCAUCAUGGUGAAAACUAAAAUCAAAUUGCCAC ............(((((((((.(((.(((.((........-)).))).)))((((.-...((((..((....)))))).......)))).....)))).)))))................ ( -22.20) >consensus CAAACAUUUUACGGUUUCACCUCGAAAUUAAAUUUCCCCU_UUCAAUACCGGCCAAAAAAGCCGCAUCGAUAGACGGCCAAAACGUGGCAUCAUGGUGAAAACUAAAAUCAAAUUGCCAC .....((((((...(((((((..(((((...)))))...............((((.....((((..((....)))))).......)))).....)))))))..))))))........... (-19.98 = -20.78 + 0.80)

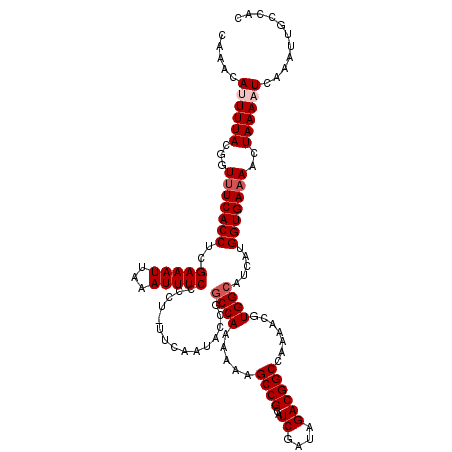
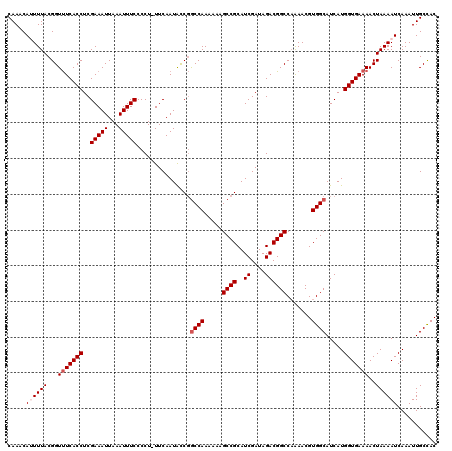
| Location | 10,514,120 – 10,514,239 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.99 |
| Mean single sequence MFE | -32.70 |
| Consensus MFE | -31.10 |
| Energy contribution | -31.18 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10514120 119 - 20766785 GUGGCAAUUUGAUUUUAGUUUUCACCAUGAUGCCACGUUUUGGCCGUCUAUCGAUGCGGCUUUUUUGGUCGGUAUUGAA-AGGGGAAAUUUAAUUUCGAGGUGAAACCGUAAAAUGUUUG ...........(((((((.(((((((.(((.((((.....))))(.(((.(((((((((((.....)))).))))))).-))).)..........))).))))))).).))))))..... ( -31.20) >DroSec_CAF1 14706 120 - 1 GUAGCAAUUUGAUUUUAGUUUUCACCAUGAUGCCACGUUUUGGCCGUCUAUCGAUGCGGCUUUUUUGGCCGGUAUUGAAAAGGGGAAAUUUAAUUUCGAGGUGAAACCGUAAAAUGUUUG ((..((.(((((..((((.((((.((..(((((((.....))).))))..(((((((((((.....)))).)))))))...)).)))).))))..))))).))..))............. ( -32.20) >DroSim_CAF1 9727 120 - 1 GUGGCAAUUUGAUUUUAGUUUUCACCAUGAUGCCACGUUUUGGCCGUCUAUCGAUGCGGCUUUUUUGGCCGGUAUUGAAAAGGGGAAAUUUAAUUUCGAGGUGAAACCGUAAAAUGUUUG ((..((.(((((..((((.((((.((..(((((((.....))).))))..(((((((((((.....)))).)))))))...)).)))).))))..))))).))..))............. ( -33.20) >DroEre_CAF1 14852 119 - 1 GUGGCAAUUUGAUUUUAGUUUUCACCAUGAUGCCACGUUUUGGCCGUCUAUCGAUGCGGCUUUUUUGGCGGGCAUUGAA-AGGGGAAAUUUAAUUUCGAGGUGAGACCGUAAAGUGUUUG ((..((.(((((..((((.((((.((.(((((((.((((..((((((........)))))).....)))))))))))..-..)))))).))))..))))).))..))............. ( -33.80) >DroYak_CAF1 11418 118 - 1 GUGGCAAUUUGAUUUUAGUUUUCACCAUGAUGCCACGUUCGGGCCGUCUAUCGAUGCGGCUUU-UUGGCCGAUAUUGAA-AGGGGAAAUUUAAUUUCGAGGUGAAACCGUAAAGUGUUUG ...........(((((((.(((((((.(((.(((.(....))))(.(((.(((((((((((..-..))))).)))))).-))).)..........))).))))))).).))))))..... ( -33.10) >consensus GUGGCAAUUUGAUUUUAGUUUUCACCAUGAUGCCACGUUUUGGCCGUCUAUCGAUGCGGCUUUUUUGGCCGGUAUUGAA_AGGGGAAAUUUAAUUUCGAGGUGAAACCGUAAAAUGUUUG ((..((.(((((..((((.((((.((..(((((((.....))).))))..(((((((((((.....))))).))))))...)).)))).))))..))))).))..))............. (-31.10 = -31.18 + 0.08)

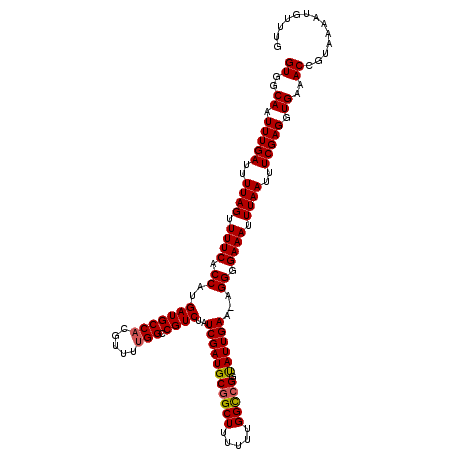
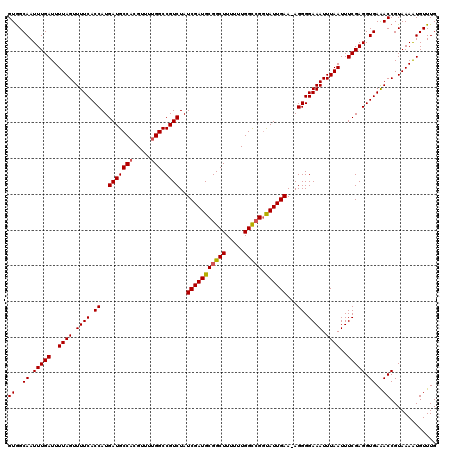
| Location | 10,514,160 – 10,514,279 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.82 |
| Mean single sequence MFE | -34.74 |
| Consensus MFE | -31.22 |
| Energy contribution | -32.06 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.75 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.39 |
| SVM RNA-class probability | 0.999887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10514160 119 + 20766785 -UUCAAUACCGACCAAAAAAGCCGCAUCGAUAGACGGCCAAAACGUGGCAUCAUGGUGAAAACUAAAAUCAAAUUGCCACGGCCAGCCAUGAAAAUCGUAGAAAUUAUCAGAUGCCAAAG -......................((((((((((..(((.....(((((((...((((..........))))...)))))))....)))(((.....))).....))))).)))))..... ( -29.50) >DroSec_CAF1 14746 120 + 1 UUUCAAUACCGGCCAAAAAAGCCGCAUCGAUAGACGGCCAAAACGUGGCAUCAUGGUGAAAACUAAAAUCAAAUUGCUACGGCCAGCCAUGAAAAUCGUGGAAAUUAUCAGAUGCCAAAG ..........(((.......)))((((((((((..((((.....((((((...((((..........))))...))))))))))..(((((.....)))))...))))).)))))..... ( -35.90) >DroSim_CAF1 9767 120 + 1 UUUCAAUACCGGCCAAAAAAGCCGCAUCGAUAGACGGCCAAAACGUGGCAUCAUGGUGAAAACUAAAAUCAAAUUGCCACGGCCAGCCAUGAAAAUCGUGGAAAUUAUCAGAUGCCAAAG ..........(((.......)))((((((((((..((((.....((((((...((((..........))))...))))))))))..(((((.....)))))...))))).)))))..... ( -38.20) >DroEre_CAF1 14892 119 + 1 -UUCAAUGCCCGCCAAAAAAGCCGCAUCGAUAGACGGCCAAAACGUGGCAUCAUGGUGAAAACUAAAAUCAAAUUGCCACGGCCAGCCAUGAAAAUCGUGAAAAUUAUCAGAUGCCAAAG -......................((((((((((..((((.....((((((...((((..........))))...))))))))))...((((.....))))....))))).)))))..... ( -31.20) >DroYak_CAF1 11458 118 + 1 -UUCAAUAUCGGCCAA-AAAGCCGCAUCGAUAGACGGCCCGAACGUGGCAUCAUGGUGAAAACUAAAAUCAAAUUGCCACGGCCAGCGAUGAAAAUCGUGGAAAUUAUCAGAUGCCAAAG -.........(((...-...)))((((((((((..((((.....((((((...((((..........))))...)))))))))).(((((....))))).....))))).)))))..... ( -38.90) >consensus _UUCAAUACCGGCCAAAAAAGCCGCAUCGAUAGACGGCCAAAACGUGGCAUCAUGGUGAAAACUAAAAUCAAAUUGCCACGGCCAGCCAUGAAAAUCGUGGAAAUUAUCAGAUGCCAAAG ..........(((.......)))((((((((((..((((.....((((((...((((..........))))...))))))))))..(((((.....)))))...))))).)))))..... (-31.22 = -32.06 + 0.84)

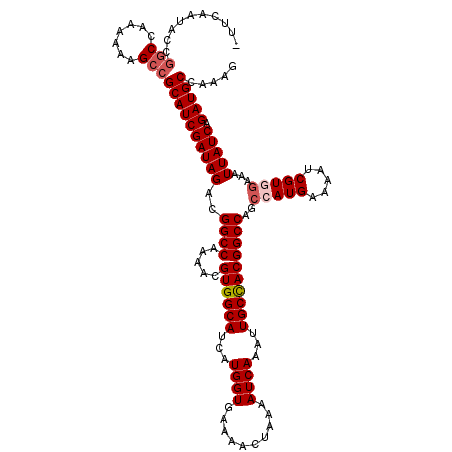
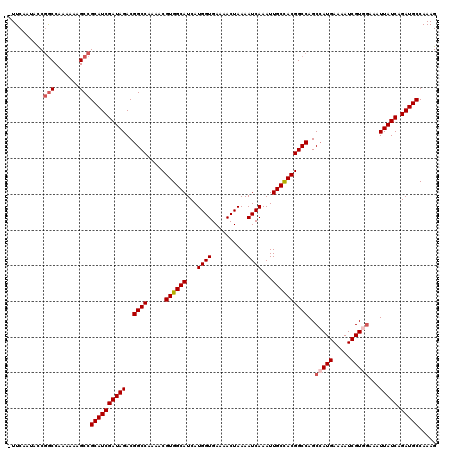
| Location | 10,514,160 – 10,514,279 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.82 |
| Mean single sequence MFE | -33.26 |
| Consensus MFE | -30.96 |
| Energy contribution | -31.08 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.980068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10514160 119 - 20766785 CUUUGGCAUCUGAUAAUUUCUACGAUUUUCAUGGCUGGCCGUGGCAAUUUGAUUUUAGUUUUCACCAUGAUGCCACGUUUUGGCCGUCUAUCGAUGCGGCUUUUUUGGUCGGUAUUGAA- ....(((...(((.((((.....)))).))).(((..(.(((((((...((..............))...)))))))..)..))))))..(((((((((((.....)))).))))))).- ( -32.24) >DroSec_CAF1 14746 120 - 1 CUUUGGCAUCUGAUAAUUUCCACGAUUUUCAUGGCUGGCCGUAGCAAUUUGAUUUUAGUUUUCACCAUGAUGCCACGUUUUGGCCGUCUAUCGAUGCGGCUUUUUUGGCCGGUAUUGAAA ...((((((((((.((((....(((.((.(((((....)))).).)).))).....)))).)))....)))))))...............(((((((((((.....)))).))))))).. ( -28.90) >DroSim_CAF1 9767 120 - 1 CUUUGGCAUCUGAUAAUUUCCACGAUUUUCAUGGCUGGCCGUGGCAAUUUGAUUUUAGUUUUCACCAUGAUGCCACGUUUUGGCCGUCUAUCGAUGCGGCUUUUUUGGCCGGUAUUGAAA ....(((...(((.((((.....)))).))).(((..(.(((((((...((..............))...)))))))..)..))))))..(((((((((((.....)))).))))))).. ( -34.94) >DroEre_CAF1 14892 119 - 1 CUUUGGCAUCUGAUAAUUUUCACGAUUUUCAUGGCUGGCCGUGGCAAUUUGAUUUUAGUUUUCACCAUGAUGCCACGUUUUGGCCGUCUAUCGAUGCGGCUUUUUUGGCGGGCAUUGAA- ...(((....(((.((((.(((.((((.((((((....)))))).)))))))....)))).)))))).((((((.((((..((((((........)))))).....))))))))))...- ( -34.50) >DroYak_CAF1 11458 118 - 1 CUUUGGCAUCUGAUAAUUUCCACGAUUUUCAUCGCUGGCCGUGGCAAUUUGAUUUUAGUUUUCACCAUGAUGCCACGUUCGGGCCGUCUAUCGAUGCGGCUUU-UUGGCCGAUAUUGAA- ....(((...............((((....))))(((..(((((((...((..............))...)))))))..)))))).....(((((((((((..-..))))).)))))).- ( -35.74) >consensus CUUUGGCAUCUGAUAAUUUCCACGAUUUUCAUGGCUGGCCGUGGCAAUUUGAUUUUAGUUUUCACCAUGAUGCCACGUUUUGGCCGUCUAUCGAUGCGGCUUUUUUGGCCGGUAUUGAA_ ....(((...(((.((((.....)))).))).(((..(.(((((((...((..............))...)))))))..)..))))))..(((((((((((.....))))).)))))).. (-30.96 = -31.08 + 0.12)

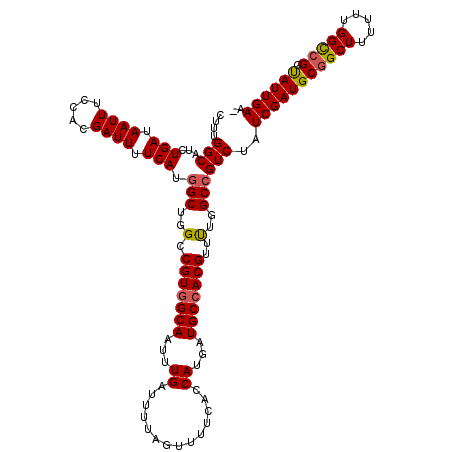
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:07 2006