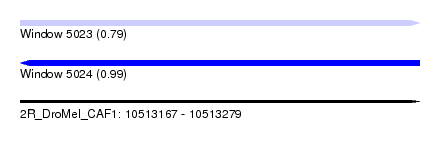
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,513,167 – 10,513,279 |
| Length | 112 |
| Max. P | 0.987611 |

| Location | 10,513,167 – 10,513,279 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 85.79 |
| Mean single sequence MFE | -20.87 |
| Consensus MFE | -16.24 |
| Energy contribution | -16.88 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10513167 112 + 20766785 AAA--AAUAAGGUCUAGUACA----A-AAAACAGAGACAGGACAACAAAAACAGGCGAGUGCCUUUGGCCAACUAUUAAAUUGGCCAGCUUAAACGUAGUCUAUAUAAUAAAUUGAUGG ...--......((((.((...----.-...))..)))).((((.((......((((....))))((((((((........)))))))).......)).))))................. ( -23.10) >DroSec_CAF1 13776 117 + 1 AAA--AAUAAGGUAUAGUACAGAAAAAAAAACAGAGACAGGACAACAAAAACAGACGAGUGCCUCUAGCCAACUAUUAAAUUGGCAAGCUUAAACGUAGUCUAUAUAAUAAAUUGAUGG ...--......((((((.((............((((.((....................)).)))).(((((........))))).............))))))))............. ( -13.75) >DroSim_CAF1 8788 116 + 1 AAA--AAUAAGGUAUAGUACAGAAAA-AAAACAGAGACAGGACAACAAAAACAGACGAGUGCCUCUGGCGAACUAUUAAAUUGGCCAGCUUAAACGUAGUCUAUAUAAUAAAUUGAUGG ...--......((((((.((......-...........(((...((............)).)))(((((.((........)).)))))..........))))))))............. ( -14.80) >DroEre_CAF1 13965 108 + 1 AAAAUAAGAAGCUGC----------A-AAAACAGAGACAGGACAACAAAAACAGGCAAGUGCCGCUGGCCAACUAUUAAAUUGGCCAGCUUAAACGUGGUCUAUAUAAUAAAUUGAUGG ...........(((.----------.-....))).....((((.((.......(((....)))(((((((((........)))))))))......)).))))................. ( -28.30) >DroYak_CAF1 10435 106 + 1 AAA--AAUAAGCUAC----------C-AAAACAGAAACAGGACAACGCAAACAGGCAAGUGCCACUGGCCAACUUUUAAAUUGGCCAGCUUAAACGUGGUCUAUACAAUAAAUUGAUGA ...--..........----------.-............((((.(((......(((....))).((((((((........))))))))......))).))))................. ( -24.40) >consensus AAA__AAUAAGGUAUAGUACA____A_AAAACAGAGACAGGACAACAAAAACAGGCGAGUGCCUCUGGCCAACUAUUAAAUUGGCCAGCUUAAACGUAGUCUAUAUAAUAAAUUGAUGG .......................................((((.((.......(((....))).((((((((........)))))))).......)).))))................. (-16.24 = -16.88 + 0.64)

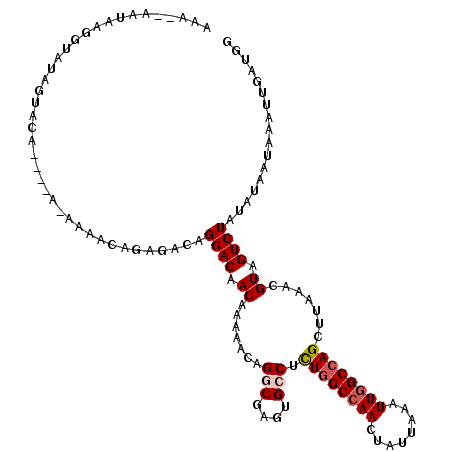
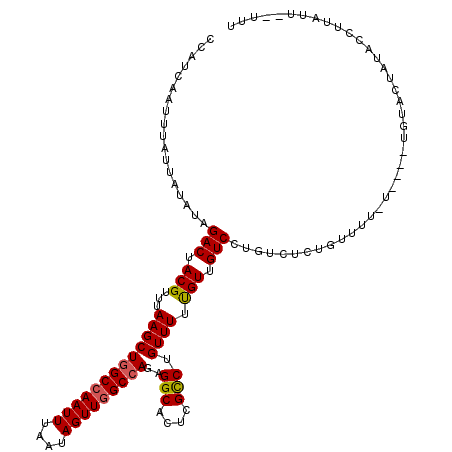
| Location | 10,513,167 – 10,513,279 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 85.79 |
| Mean single sequence MFE | -25.29 |
| Consensus MFE | -19.52 |
| Energy contribution | -19.52 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10513167 112 - 20766785 CCAUCAAUUUAUUAUAUAGACUACGUUUAAGCUGGCCAAUUUAAUAGUUGGCCAAAGGCACUCGCCUGUUUUUGUUGUCCUGUCUCUGUUUU-U----UGUACUAGACCUUAUU--UUU ..................(((.(((...(((((((((((((....))))))))).((((....)))))))).))).)))..((((..((...-.----...)).))))......--... ( -26.40) >DroSec_CAF1 13776 117 - 1 CCAUCAAUUUAUUAUAUAGACUACGUUUAAGCUUGCCAAUUUAAUAGUUGGCUAGAGGCACUCGUCUGUUUUUGUUGUCCUGUCUCUGUUUUUUUUUCUGUACUAUACCUUAUU--UUU .........(((..((((((..............(((((((....)))))))((((((((..((.(.......).))...))))))))........))))))..))).......--... ( -20.80) >DroSim_CAF1 8788 116 - 1 CCAUCAAUUUAUUAUAUAGACUACGUUUAAGCUGGCCAAUUUAAUAGUUCGCCAGAGGCACUCGUCUGUUUUUGUUGUCCUGUCUCUGUUUU-UUUUCUGUACUAUACCUUAUU--UUU ..............(((((((.......((((.(((.((((....)))).)))(((((((..((.(.......).))...))))))))))).-......)).))))).......--... ( -18.04) >DroEre_CAF1 13965 108 - 1 CCAUCAAUUUAUUAUAUAGACCACGUUUAAGCUGGCCAAUUUAAUAGUUGGCCAGCGGCACUUGCCUGUUUUUGUUGUCCUGUCUCUGUUUU-U----------GCAGCUUCUUAUUUU ..................(((.(((.....(((((((((((....)))))))))))(((....)))......))).)))......((((...-.----------))))........... ( -30.30) >DroYak_CAF1 10435 106 - 1 UCAUCAAUUUAUUGUAUAGACCACGUUUAAGCUGGCCAAUUUAAAAGUUGGCCAGUGGCACUUGCCUGUUUGCGUUGUCCUGUUUCUGUUUU-G----------GUAGCUUAUU--UUU ..(((((..((....((((((.((((....(((((((((((....)))))))))))(((....))).....)))).)).))))...))..))-)----------))........--... ( -30.90) >consensus CCAUCAAUUUAUUAUAUAGACUACGUUUAAGCUGGCCAAUUUAAUAGUUGGCCAGAGGCACUCGCCUGUUUUUGUUGUCCUGUCUCUGUUUU_U____UGUACUAUACCUUAUU__UUU ..................(((.(((...(((((((((((((....)))))))))..(((....))).)))).))).)))........................................ (-19.52 = -19.52 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:46:03 2006