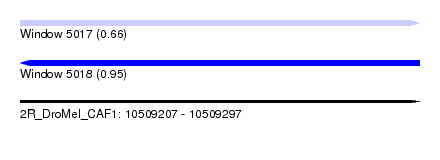
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,509,207 – 10,509,297 |
| Length | 90 |
| Max. P | 0.952336 |

| Location | 10,509,207 – 10,509,297 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 94.78 |
| Mean single sequence MFE | -15.40 |
| Consensus MFE | -13.73 |
| Energy contribution | -13.97 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10509207 90 + 20766785 AUGCAAUUGAAAUUAAAUCGCUUGCAAAUGAUUCGCAAGCUACCUAAAAAAAAAUUGGGCAAUCCAAAAAAUGACAUCGUUUAUGCAGCC .((((((((..........((((((.........))))))..(((((.......))))))))).....(((((....))))).))))... ( -17.60) >DroSec_CAF1 9859 89 + 1 AUGCAAUUGAAAUUAAAUCGCUUGCAAAUGAUUCGCAAGCCACCUAAAA-AAAAUUGGGCAAUCCAAAAAAUUACAUCGUUUAUGCAGCC .((((..(((.........((((((.........))))))..(((((..-....))))).................)))....))))... ( -15.50) >DroEre_CAF1 10047 88 + 1 AUGCAAUUGAAAUUAAAUCGCCUGCAAAUGAUUCGCAAGCCACCUAAAA--AAAUUGGCCAAUCCAAAAAAUGACAUCGUUUAUGCAGCC .((((..(((.......)))..))))........(((.((((.......--....)))).........(((((....))))).))).... ( -12.60) >DroYak_CAF1 6587 88 + 1 AUGCAAUUGAAAUUAAAUCGCUUGCAAAUGAUUCGCAAGCCACCUAAAA--AAAUUGGGAAAUCCAAAAAAUGACAUCGCUUAUGCAGGC .((((..............((((((.........))))))..(((((..--...)))))........................))))... ( -15.90) >consensus AUGCAAUUGAAAUUAAAUCGCUUGCAAAUGAUUCGCAAGCCACCUAAAA__AAAUUGGGCAAUCCAAAAAAUGACAUCGUUUAUGCAGCC .((((..............((((((.........))))))..............((((.....))))................))))... (-13.73 = -13.97 + 0.25)


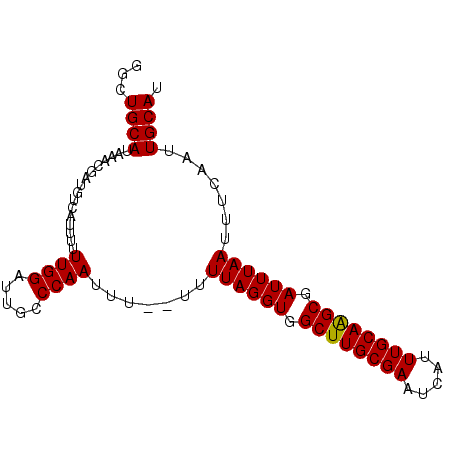
| Location | 10,509,207 – 10,509,297 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 94.78 |
| Mean single sequence MFE | -21.35 |
| Consensus MFE | -20.84 |
| Energy contribution | -20.65 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10509207 90 - 20766785 GGCUGCAUAAACGAUGUCAUUUUUUGGAUUGCCCAAUUUUUUUUUAGGUAGCUUGCGAAUCAUUUGCAAGCGAUUUAAUUUCAAUUGCAU ...((((................((((.....)))).......((((((.((((((((.....)))))))).)))))).......)))). ( -19.20) >DroSec_CAF1 9859 89 - 1 GGCUGCAUAAACGAUGUAAUUUUUUGGAUUGCCCAAUUUU-UUUUAGGUGGCUUGCGAAUCAUUUGCAAGCGAUUUAAUUUCAAUUGCAU .............((((((((..((((.....))))....-..((((((.((((((((.....)))))))).))))))....)))))))) ( -22.60) >DroEre_CAF1 10047 88 - 1 GGCUGCAUAAACGAUGUCAUUUUUUGGAUUGGCCAAUUU--UUUUAGGUGGCUUGCGAAUCAUUUGCAGGCGAUUUAAUUUCAAUUGCAU ((((((((.....))))((.....))....)))).....--..((((((.((((((((.....)))))))).))))))............ ( -20.80) >DroYak_CAF1 6587 88 - 1 GCCUGCAUAAGCGAUGUCAUUUUUUGGAUUUCCCAAUUU--UUUUAGGUGGCUUGCGAAUCAUUUGCAAGCGAUUUAAUUUCAAUUGCAU ..........(((((........((((.....))))...--..((((((.((((((((.....)))))))).)))))).....))))).. ( -22.80) >consensus GGCUGCAUAAACGAUGUCAUUUUUUGGAUUGCCCAAUUU__UUUUAGGUGGCUUGCGAAUCAUUUGCAAGCGAUUUAAUUUCAAUUGCAU ...((((................((((.....)))).......((((((.((((((((.....)))))))).)))))).......)))). (-20.84 = -20.65 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:57 2006