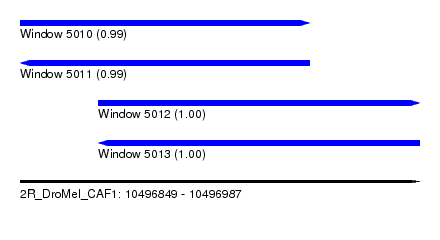
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,496,849 – 10,496,987 |
| Length | 138 |
| Max. P | 0.999843 |

| Location | 10,496,849 – 10,496,949 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.34 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -22.88 |
| Energy contribution | -24.28 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10496849 100 + 20766785 CUAUCACCUGCUCUUCUUUGCAGUGCAGUUUUUGUUGGUACAGUGCAUACCCGAUUCGGUGAAUUGUGAAGACCGAG--GGUAUCCACUGUACCAAAGUGAU ..(((((((((.((.......)).))))......(((((((((((.((((((...(((((...........))))))--))))).))))))))))).))))) ( -40.80) >DroSec_CAF1 10210 100 + 1 CUAUCACCUGUUCUUCUUUGCAGUGCAGUUUUUGUUGUUACAGUGCUUACUCGAUUCGGUGAAUUGUGUAGACCGAG--GGUGUUCACUGUACCAAAGUGAU ..(((((((((........))))...........(((.(((((((..(((((...(((((...........))))))--))))..))))))).))).))))) ( -27.80) >DroSim_CAF1 10178 100 + 1 CUAUCACCUGUUCUUCUUUGCAGUGCAGUUUUUGUUGGUACAGUGAUUACUCGAUUCGGUGAAUUGUAUAGACCGAG--GGUGUUUACUGUACCAAAGUGAU ..(((((((((........))))...........((((((((((((.(((((...(((((...........))))))--)))).)))))))))))).))))) ( -33.70) >DroEre_CAF1 10403 99 + 1 CUAUCGGCUGUUCUCCUAUACAGUGCAGUUUUUGUUGGUACAGUGCUUACUCGGUACGGUGAAUUGUGAAGCCCAAGGGGGUGUUCACUGUACCAAAG---U ......(((((...((...((((........)))).)).)))))........((((((((((((......((((....))))))))))))))))....---. ( -32.20) >DroYak_CAF1 10523 86 + 1 CUAUCGCCUGUUCUGCUAUGCAGUGCAGUUUUU-UUGGUACAGUGCUUACCGGCUAGGGUGAAUUG---------------UGUUCACUGUACCAAAUUUAU .......((((.((((...)))).))))....(-(((((((((((.(((((......)))))....---------------....))))))))))))..... ( -28.00) >consensus CUAUCACCUGUUCUUCUUUGCAGUGCAGUUUUUGUUGGUACAGUGCUUACUCGAUUCGGUGAAUUGUGAAGACCGAG__GGUGUUCACUGUACCAAAGUGAU ....((.((((........)))))).........(((((((((((..((((...((((((...........))))))..))))..)))))))))))...... (-22.88 = -24.28 + 1.40)

| Location | 10,496,849 – 10,496,949 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.34 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -16.55 |
| Energy contribution | -17.83 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.54 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10496849 100 - 20766785 AUCACUUUGGUACAGUGGAUACC--CUCGGUCUUCACAAUUCACCGAAUCGGGUAUGCACUGUACCAACAAAAACUGCACUGCAAAGAAGAGCAGGUGAUAG (((((.(((((((((((.(((((--((((((...........)))))...)))))).)))))))))))......((((.((.......)).))))))))).. ( -44.20) >DroSec_CAF1 10210 100 - 1 AUCACUUUGGUACAGUGAACACC--CUCGGUCUACACAAUUCACCGAAUCGAGUAAGCACUGUAACAACAAAAACUGCACUGCAAAGAAGAACAGGUGAUAG (((((((((.(((((((......--(((((((.............).))))))....)))))))....))))..(((..((.......))..)))))))).. ( -22.12) >DroSim_CAF1 10178 100 - 1 AUCACUUUGGUACAGUAAACACC--CUCGGUCUAUACAAUUCACCGAAUCGAGUAAUCACUGUACCAACAAAAACUGCACUGCAAAGAAGAACAGGUGAUAG (((((.((((((((((.......--.(((((...........)))))...((....))))))))))))......(((..((.......))..)))))))).. ( -27.10) >DroEre_CAF1 10403 99 - 1 A---CUUUGGUACAGUGAACACCCCCUUGGGCUUCACAAUUCACCGUACCGAGUAAGCACUGUACCAACAAAAACUGCACUGUAUAGGAGAACAGCCGAUAG .---..(((((((((((....(((....))).......((((........))))...)))))))))))...........((((...((.......)).)))) ( -23.40) >DroYak_CAF1 10523 86 - 1 AUAAAUUUGGUACAGUGAACA---------------CAAUUCACCCUAGCCGGUAAGCACUGUACCAA-AAAAACUGCACUGCAUAGCAGAACAGGCGAUAG .....((((((((((((....---------------......(((......)))...)))))))))))-)....(((..((((...))))..)))....... ( -24.32) >consensus AUCACUUUGGUACAGUGAACACC__CUCGGUCUACACAAUUCACCGAAUCGAGUAAGCACUGUACCAACAAAAACUGCACUGCAAAGAAGAACAGGUGAUAG ......(((((((((((.........((((.............))))..........)))))))))))......(((..((.......))..)))....... (-16.55 = -17.83 + 1.28)

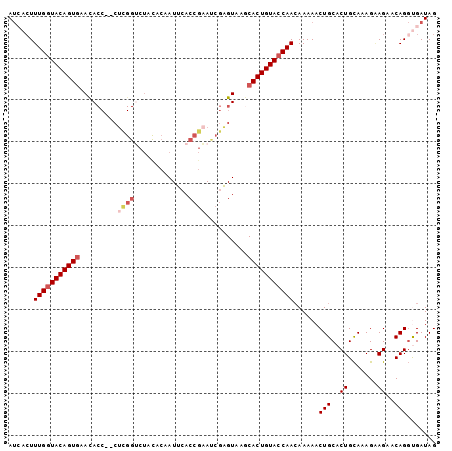
| Location | 10,496,876 – 10,496,987 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 75.38 |
| Mean single sequence MFE | -31.24 |
| Consensus MFE | -19.22 |
| Energy contribution | -21.18 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.62 |
| SVM decision value | 3.83 |
| SVM RNA-class probability | 0.999644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10496876 111 + 20766785 GUUUUUGUUGGUACAGUGCAUACCCGAUUCGGUGAAUUGUGAAGACCGAG--GGUAUCCACUGUACCAAAGUGAUAAAUGUGCACCUAGAUUUUCAUAC-AAAAA-UAUGUUUAC ((((((((((((((((((.((((((...(((((...........))))))--))))).))))))))))..((((.((((.((....)).))))))))))-)))))-)........ ( -38.00) >DroSec_CAF1 10237 112 + 1 GUUUUUGUUGUUACAGUGCUUACUCGAUUCGGUGAAUUGUGUAGACCGAG--GGUGUUCACUGUACCAAAGUGAUAAUUGUGCCCCCAGAUUUUCAUAC-AAAAAUUAUGUUUAC ((((((((((.(((((((..(((((...(((((...........))))))--))))..))))))).))..((((.((((.........)))).))))))-))))))......... ( -26.90) >DroSim_CAF1 10205 112 + 1 GUUUUUGUUGGUACAGUGAUUACUCGAUUCGGUGAAUUGUAUAGACCGAG--GGUGUUUACUGUACCAAAGUGAUAAUUGUGCACCUAGAUUUUCAUAC-AAAAAUUAUGUUUAC (((((((((((((((((((.(((((...(((((...........))))))--)))).)))))))))))..((((.((((.........)))).))))))-))))))......... ( -32.80) >DroEre_CAF1 10430 99 + 1 GUUUUUGUUGGUACAGUGCUUACUCGGUACGGUGAAUUGUGAAGCCCAAGGGGGUGUUCACUGUACCAAAG---UAAAUCUG----------UUCAUCAAAAA---UGUGUUAAC (((((((.(((.((((...(((((.((((((((((((......((((....))))))))))))))))..))---)))..)))----------)))).))))))---)........ ( -35.90) >DroYak_CAF1 10550 90 + 1 GUUUUU-UUGGUACAGUGCUUACCGGCUAGGGUGAAUUG---------------UGUUCACUGUACCAAAUUUAUAAAUGUGA--UUUGAUUUUCAUGU-AAA---UCUCUU--- .....(-(((((((((((.(((((......)))))....---------------....)))))))))))).......((((((--........))))))-...---......--- ( -22.60) >consensus GUUUUUGUUGGUACAGUGCUUACUCGAUUCGGUGAAUUGUGAAGACCGAG__GGUGUUCACUGUACCAAAGUGAUAAAUGUGC_CCUAGAUUUUCAUAC_AAAAA_UAUGUUUAC .......(((((((((((..((((...((((((...........))))))..))))..))))))))))).............................................. (-19.22 = -21.18 + 1.96)

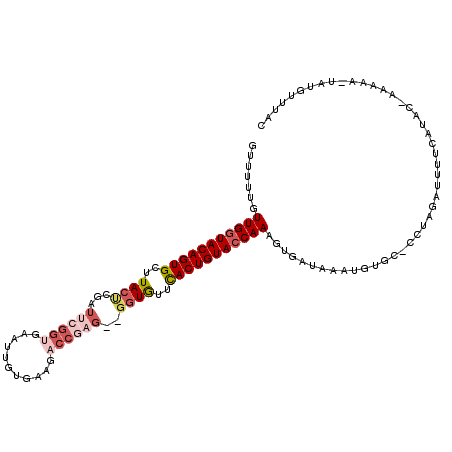

| Location | 10,496,876 – 10,496,987 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 75.38 |
| Mean single sequence MFE | -25.43 |
| Consensus MFE | -14.67 |
| Energy contribution | -15.95 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.58 |
| SVM decision value | 4.23 |
| SVM RNA-class probability | 0.999843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10496876 111 - 20766785 GUAAACAUA-UUUUU-GUAUGAAAAUCUAGGUGCACAUUUAUCACUUUGGUACAGUGGAUACC--CUCGGUCUUCACAAUUCACCGAAUCGGGUAUGCACUGUACCAACAAAAAC .........-(((((-((..........(((((.........)))))((((((((((.(((((--((((((...........)))))...)))))).))))))))))))))))). ( -37.40) >DroSec_CAF1 10237 112 - 1 GUAAACAUAAUUUUU-GUAUGAAAAUCUGGGGGCACAAUUAUCACUUUGGUACAGUGAACACC--CUCGGUCUACACAAUUCACCGAAUCGAGUAAGCACUGUAACAACAAAAAC ..........(((((-((.((.....(..(((............)))..)(((((((......--(((((((.............).))))))....))))))).))))))))). ( -20.02) >DroSim_CAF1 10205 112 - 1 GUAAACAUAAUUUUU-GUAUGAAAAUCUAGGUGCACAAUUAUCACUUUGGUACAGUAAACACC--CUCGGUCUAUACAAUUCACCGAAUCGAGUAAUCACUGUACCAACAAAAAC ..........(((((-((..........(((((.........)))))(((((((((.......--.(((((...........)))))...((....)))))))))))))))))). ( -24.50) >DroEre_CAF1 10430 99 - 1 GUUAACACA---UUUUUGAUGAA----------CAGAUUUA---CUUUGGUACAGUGAACACCCCCUUGGGCUUCACAAUUCACCGUACCGAGUAAGCACUGUACCAACAAAAAC .........---((((((.((.(----------(((.((((---(((.(((((.(((((..(((....)))........))))).))))))))))))..))))..)).)))))). ( -25.30) >DroYak_CAF1 10550 90 - 1 ---AAGAGA---UUU-ACAUGAAAAUCAAA--UCACAUUUAUAAAUUUGGUACAGUGAACA---------------CAAUUCACCCUAGCCGGUAAGCACUGUACCAA-AAAAAC ---....((---(((-......)))))...--.............((((((((((((....---------------......(((......)))...)))))))))))-)..... ( -19.92) >consensus GUAAACAUA_UUUUU_GUAUGAAAAUCUAGG_GCACAUUUAUCACUUUGGUACAGUGAACACC__CUCGGUCUACACAAUUCACCGAAUCGAGUAAGCACUGUACCAACAAAAAC ..............................................(((((((((((.........((((.............))))..........)))))))))))....... (-14.67 = -15.95 + 1.28)

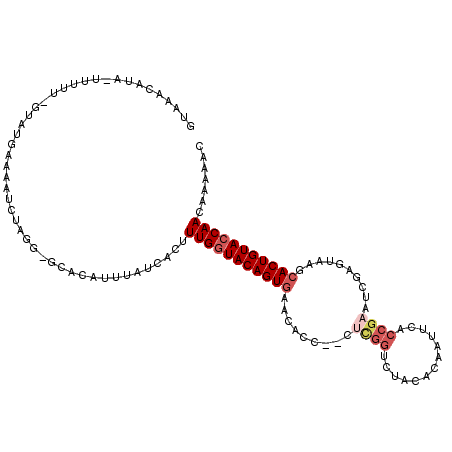
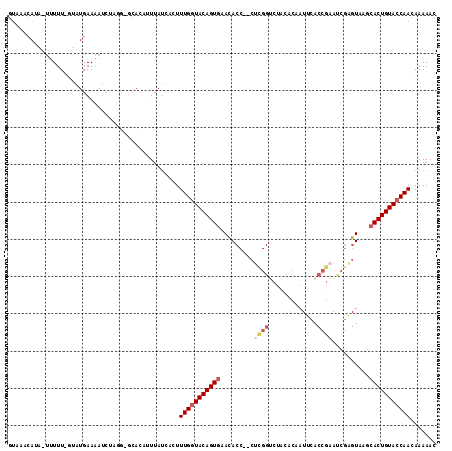
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:53 2006