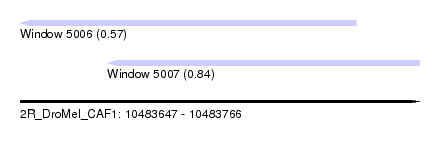
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,483,647 – 10,483,766 |
| Length | 119 |
| Max. P | 0.844043 |

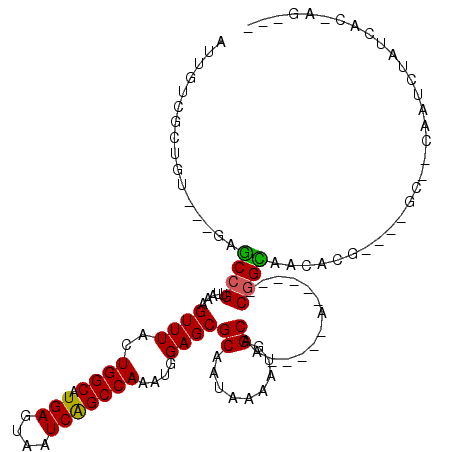
| Location | 10,483,647 – 10,483,747 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.48 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -17.26 |
| Energy contribution | -16.90 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10483647 100 - 20766785 UUUGUCGCUGUCGAGAGCCGUUAAAGUUUACUGGCAUGAGUAAUCAGCCAAAUGGAGCGCAAUAAAAUGGCAA-----A------CGGUAACACG----GC--CAAUCUAUCACCAG--- ......(((((.(...((((((...((((..((((.(((....)))))))....))))((.........)).)-----)------))))..))))----))--..............--- ( -24.70) >DroVir_CAF1 114394 99 - 1 AUCGUCGCUGU---GCGCCGUUAAAGUUUACUGGCAUGAGUAAUCAGCCAAAUGGAGCGCAAUAAAAUGGCAACGGCAA------CGGCAACACG----GC--CAAUCUAUCAG------ ...((.(((((---..((((((...((((..((((.(((....)))))))....)))).......)))))).))))).)------)(((......----))--)..........------ ( -31.50) >DroPse_CAF1 81400 98 - 1 AUUGUAUCGG--GAGAACCGUUAAAGUUUACUGGCAUGAGUAAUCGGCCAAAUGGAGCGCAAUAAAAUGGCAA-----A------CGGUAACACG----GC--CAAUAUAUCACAAG--- (((((..(((--.....))).....((((..((((.(((....)))))))....)))))))))....((((..-----.------((......))----))--))............--- ( -22.40) >DroGri_CAF1 102754 107 - 1 GUUGUC----------GCCGUUAAAGUUUACUGGCAUGAGUAAUCAGCCAAAUGGAGCGCAAUAAAAUGGCAACGGCAACGGCAACAGCAACACAGGCGGCAACAAUCUAUCAGCAG--- ((((((----------(((......((((..((((.(((....)))))))....))))((........(....)(....)(....).))......))))))))).............--- ( -34.80) >DroMoj_CAF1 136744 104 - 1 GUUGUCGCGGU---GAGCCGUUAAAGUUUACUGGCAUGAGUAAUCAGCCAAAUGGAGCGCAAUAAAAUGGCAACGGCAACGG-AACGGCAACACG----GC--CAAUCUAUCAG------ ((((((((.((---..((((((...((((..((((.(((....)))))))....))))((.........)))))))).)).)-..)))))))...----..--...........------ ( -32.40) >DroAna_CAF1 90850 103 - 1 UCUGUCGCUGUCGAGGGCCGUUAAAGUUUACUGGCAUGAGUAAUCAGCCAAAUGGAGCGCAAUAAAAUGGCAA-----A------CGGUAACACG----GC--CAAUCUAUCACACAAGU ......(((((.(...((((((...((((..((((.(((....)))))))....))))((.........)).)-----)------))))..))))----))--................. ( -25.80) >consensus AUUGUCGCUGU___GAGCCGUUAAAGUUUACUGGCAUGAGUAAUCAGCCAAAUGGAGCGCAAUAAAAUGGCAA_____A______CGGCAACACG____GC__CAAUCUAUCAC_AG___ ................((((.....((((..((((.(((....)))))))....))))((.........))..............))))............................... (-17.26 = -16.90 + -0.36)


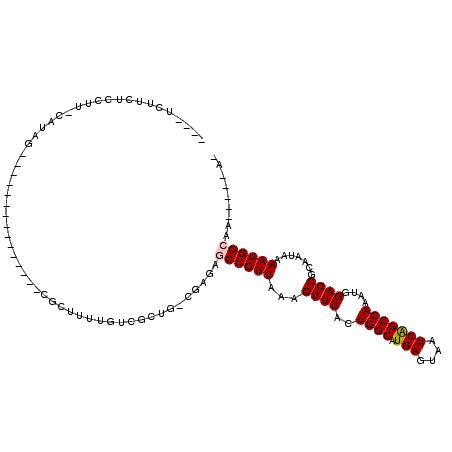
| Location | 10,483,673 – 10,483,766 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.29 |
| Mean single sequence MFE | -24.98 |
| Consensus MFE | -15.56 |
| Energy contribution | -15.67 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10483673 93 - 20766785 ----UCUUUUCUUU-CAUAG---------------CGUUUUUGUCGCUGUCGAGAGCCGUUAAAGUUUACUGGCAUGAGUAAUCAGCCAAAUGGAGCGCAAUAAAAUGGCAA-----A- ----.......(((-(((((---------------((.......)))))).))))((((((...((((..((((.(((....)))))))....)))).......))))))..-----.- ( -26.80) >DroPse_CAF1 81426 92 - 1 ----UCCCCCCCUUUUUUCC---------------CACUAUUGUAUCGG--GAGAACCGUUAAAGUUUACUGGCAUGAGUAAUCGGCCAAAUGGAGCGCAAUAAAAUGGCAA-----A- ----...............(---------------((.((((((..(((--.....))).....((((..((((.(((....)))))))....))))))))))...)))...-----.- ( -19.90) >DroGri_CAF1 102791 92 - 1 GUUAAAUUGUCAAU-CA-AU---------------UGCUGUUGUC----------GCCGUUAAAGUUUACUGGCAUGAGUAAUCAGCCAAAUGGAGCGCAAUAAAAUGGCAACGGCAAC ..............-..-.(---------------((((((((((----------(...(((..((((..((((.(((....)))))))....))))....)))..)))))))))))). ( -27.50) >DroYak_CAF1 88407 93 - 1 ----UUUUCUCUUU-CAUAG---------------CGUUUUUGUCGCUGUCGAGAGCCGUUAAAGUUUACUGGCAUGAGUAAUCAGCCAAAUGGAGCGCAAUAAAAUGGCAA-----A- ----...((((...-.((((---------------((.......)))))).))))((((((...((((..((((.(((....)))))))....)))).......))))))..-----.- ( -27.20) >DroAna_CAF1 90879 107 - 1 ----UCUUCGG-UU-UAUAGUUUCUGACUCUCUUUAGUCUCUGUCGCUGUCGAGGGCCGUUAAAGUUUACUGGCAUGAGUAAUCAGCCAAAUGGAGCGCAAUAAAAUGGCAA-----A- ----.(((((.-..-.(((((..(.((((......))))...)..))))))))))((((((...((((..((((.(((....)))))))....)))).......))))))..-----.- ( -28.40) >DroPer_CAF1 87947 92 - 1 ----UCCCCCCCUU-UUUCC---------------CACUAUUGUAUCGG-GGAGAACCGUUAAAGUUUACUGGCAUGAGUAAUCGGCCAAAUGGAGCGCAAUAAAAUGGCAA-----A- ----..........-....(---------------((.((((((..(((-......))).....((((..((((.(((....)))))))....))))))))))...)))...-----.- ( -20.10) >consensus ____UCUUCUCCUU_CAUAG_______________CGCUUUUGUCGCUG_CGAGAGCCGUUAAAGUUUACUGGCAUGAGUAAUCAGCCAAAUGGAGCGCAAUAAAAUGGCAA_____A_ .......................................................((((((...((((..((((.(((....)))))))....)))).......))))))......... (-15.56 = -15.67 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:47 2006