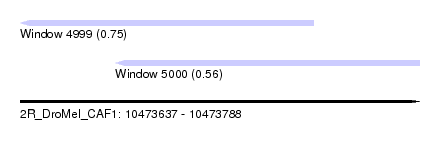
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,473,637 – 10,473,788 |
| Length | 151 |
| Max. P | 0.751019 |

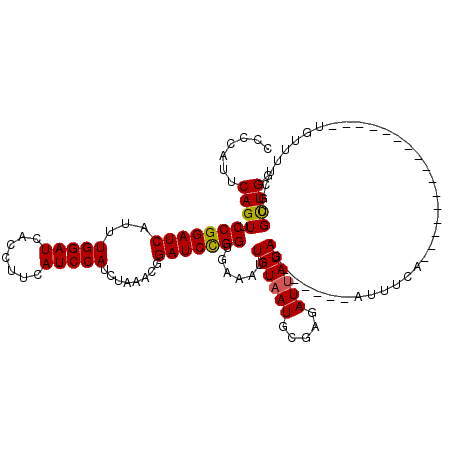
| Location | 10,473,637 – 10,473,748 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.56 |
| Mean single sequence MFE | -23.78 |
| Consensus MFE | -15.64 |
| Energy contribution | -15.53 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10473637 111 - 20766785 CCCCACUCAGUUCCGGAUCAUUUGGAUCACCUUCAUCCAUCGAAACGGAUCCGG---AAUUGUAAUUUCAGAUUAUAG------AAUUCUUACUUUACUCUUACUCUGUUUACGUGUUGG .((.((.((((((((((((...(((((.......)))))........)))))))---))))((((...((((.((.((------(.............))))).)))).))))).)).)) ( -27.82) >DroSec_CAF1 77147 98 - 1 CCACAUUCAGUUCCGGAUCAUUUGGAUCACCUUCAUCCAUCUAAACGGAUCCGGUGAAAUUGUUAUGCGAGAUUAUAG------AUUUCA----------------UGUUUUCGUGCUGG .......((((.(((((((.(((((((...........)))))))..)))))))............((((((.(((.(------....))----------------)).)))))))))). ( -23.20) >DroSim_CAF1 77691 104 - 1 CCCCAUUCAGUUCCGGAUCAUUUGGAUCACCUUCAUCCAUCUUAACGGAUCUGGUGAAAUUGUAAUGCGAUAUUAUACUUAAGAGUUUCA----------------UGCUUUCUUGCUGG ..(((..(((.((((.......(((((.......)))))......)))).)))((((((((((((((...))))))).......))))))----------------)((......))))) ( -20.33) >consensus CCCCAUUCAGUUCCGGAUCAUUUGGAUCACCUUCAUCCAUCUAAACGGAUCCGGUGAAAUUGUAAUGCGAGAUUAUAG______AUUUCA________________UGUUUUCGUGCUGG .......((((.(((((((...(((((.......)))))........)))))))......((((((.....))))))......................................)))). (-15.64 = -15.53 + -0.11)

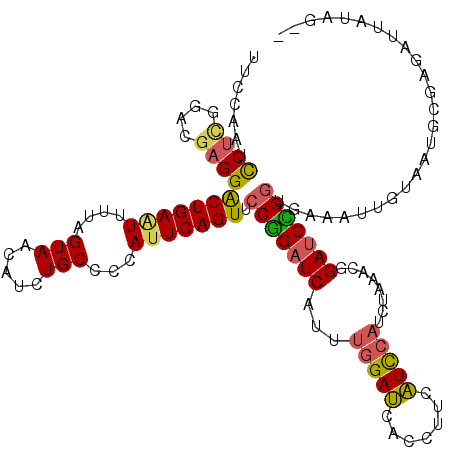
| Location | 10,473,673 – 10,473,788 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.51 |
| Mean single sequence MFE | -27.26 |
| Consensus MFE | -18.14 |
| Energy contribution | -18.20 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10473673 115 - 20766785 UUCCAAUCCUUGGACGAGGACUGAAUUUUAGUAACAUCUGCCCCACUCAGUUCCGGAUCAUUUGGAUCACCUUCAUCCAUCGAAACGGAUCCGG---AAUUGUAAUUUCAGAUUAUAG-- ...((.(((((....))))).))............(((((.......((((((((((((...(((((.......)))))........)))))))---)))))......))))).....-- ( -32.92) >DroSec_CAF1 77167 118 - 1 UUCCAAUCCUCCGACGAGGGCUGAAUUUUAGUAACAUCUGCCACAUUCAGUUCCGGAUCAUUUGGAUCACCUUCAUCCAUCUAAACGGAUCCGGUGAAAUUGUUAUGCGAGAUUAUAG-- ...((..((((....))))..))((((((.((((((.(((.......)))..(((((((.(((((((...........)))))))..)))))))......))))))..))))))....-- ( -29.30) >DroSim_CAF1 77715 120 - 1 UUCCAAUCCUCGGACGAGGGCUGAAUUUUAGUAACAUCUGCCCCAUUCAGUUCCGGAUCAUUUGGAUCACCUUCAUCCAUCUUAACGGAUCUGGUGAAAUUGUAAUGCGAUAUUAUACUU .(((((((....))((.(((((((((....(((.....)))...)))))))))))......)))))(((((...((((........))))..))))).((((.....))))......... ( -28.40) >DroYak_CAF1 78534 95 - 1 UAACAAUUCCCGGACAAGGACUGAGUUUUAGUAUCAUCUGCCCUAUUCAGUUGCAGAUCAGGAGGACAACCAUCGUU-----AAACUGA-CUGGUAUAAUU------------------- .........((((.....((((((((..(((......)))....)))))))).....((((...(((.......)))-----...))))-)))).......------------------- ( -18.40) >consensus UUCCAAUCCUCGGACGAGGACUGAAUUUUAGUAACAUCUGCCCCAUUCAGUUCCGGAUCAUUUGGAUCACCUUCAUCCAUCUAAACGGAUCCGGUGAAAUUGUAAUGCGAGAUUAUAG__ .......((((....))))(((((((....(((.....)))...))))))).(((((((...(((((.......)))))........))))))).......................... (-18.14 = -18.20 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:41 2006