
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,455,235 – 10,455,330 |
| Length | 95 |
| Max. P | 0.613535 |

| Location | 10,455,235 – 10,455,330 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.59 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -16.08 |
| Energy contribution | -15.92 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
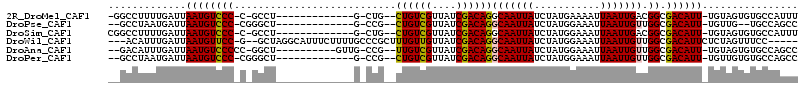
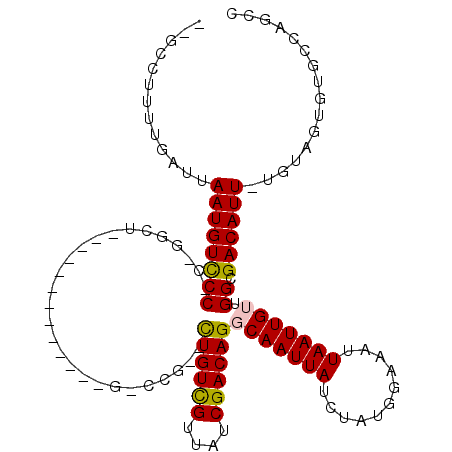
>2R_DroMel_CAF1 10455235 95 + 20766785 -GGCCUUUUGAUUAAUGUCCC-C-GCCU-------------G-CUG--CUGUCGUUAUCGACAGGCAAUUAUCUAUGAAAAUUAAUUGACGGCGACAUU-UGUAGUGUGCCAUUU -(((...(((...((((((.(-(-(...-------------.-...--((((((....)))))).((((((...........)))))).))).))))))-..)))...))).... ( -27.00) >DroPse_CAF1 54795 93 + 1 --GCCUAAUGAUUAAUGUCCC-CGGGCU-------------G-CCG--CUGUCGUUAUCGACAGGCAAUUAUCUAUGGAAAUUAAUUGUUGGCGACAUU-UGUUG--UGCCAGCC --.......(((((((.(((.-.(((.(-------------(-((.--.(((((....)))))))))....)))..))).)))))))((((((.(((..-...))--))))))). ( -29.10) >DroSim_CAF1 59601 96 + 1 CGGCCUUUUGAUUAAUGUCCC-C-GCCU-------------G-CUG--CUGUCGUUAUCGACAGGCAAUUAUCUAUGGAAAUUAAUUGACGGCGACAUU-UGUAGUGUGCCAUUU .(((...(((...((((((.(-(-(...-------------.-...--((((((....)))))).((((((...........)))))).))).))))))-..)))...))).... ( -27.30) >DroWil_CAF1 88817 104 + 1 ---ACAUUUGAUUAAUGUUCC-G--GCUAGGCAUUUCUUUUGCCCGCUUUGUUGUUAUCGACAGGCAAUUAUCUAUGGAAAUUAAUUGUUGGCGACAUUCUCUAGUUUCC----- ---......(((((((.((((-(--....((((.......)))).((((.((((....)))))))).........))))))))))))...((.(((........))).))----- ( -21.90) >DroAna_CAF1 62840 98 + 1 --GACAUUUGAUUAAUGUCCCCC-GGCU----------GUUG-CCG--UUGUCGUUAUCGACAGGCAAUUAUCUAUGGAAAUUAAUUGUUGGCGACAUU-UGUAGUGUGCCAGCC --.......(((((((.(((...-((..----------((((-((.--.(((((....)))))))))))..))...))).)))))))((((((.((((.-....)))))))))). ( -33.30) >DroPer_CAF1 61011 95 + 1 --GCCUAAUGAUUAAUGUCCC-CGGGCU-------------G-CCG--CUGUCGUUAUCGACAGGCAAUUAUCUAUGGAAAUUAAUUGUUGGCGACAUU-UGUUGUGUGCCAGCC --.......(((((((.(((.-.(((.(-------------(-((.--.(((((....)))))))))....)))..))).)))))))((((((.((((.-....)))))))))). ( -30.20) >consensus __GCCUUUUGAUUAAUGUCCC_C_GGCU_____________G_CCG__CUGUCGUUAUCGACAGGCAAUUAUCUAUGGAAAUUAAUUGUUGGCGACAUU_UGUAGUGUGCCAGCC .............((((((((...........................((((((....))))))(((((((...........))))))).)).))))))................ (-16.08 = -15.92 + -0.17)
| Location | 10,455,235 – 10,455,330 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.59 |
| Mean single sequence MFE | -25.51 |
| Consensus MFE | -13.65 |
| Energy contribution | -14.32 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
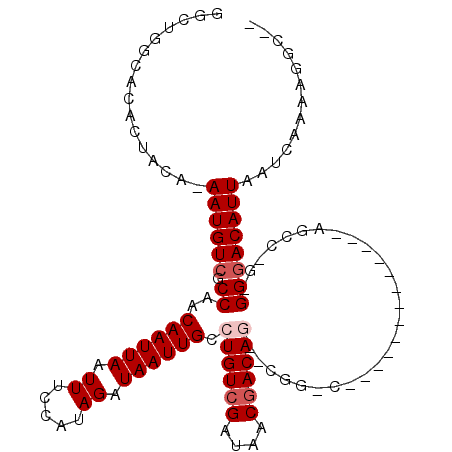
>2R_DroMel_CAF1 10455235 95 - 20766785 AAAUGGCACACUACA-AAUGUCGCCGUCAAUUAAUUUUCAUAGAUAAUUGCCUGUCGAUAACGACAG--CAG-C-------------AGGC-G-GGGACAUUAAUCAAAAGGCC- ....(((........-((((((.(((((.....((((....))))...(((((((((....))))))--..)-)-------------))))-)-).)))))).........)))- ( -29.53) >DroPse_CAF1 54795 93 - 1 GGCUGGCA--CAACA-AAUGUCGCCAACAAUUAAUUUCCAUAGAUAAUUGCCUGUCGAUAACGACAG--CGG-C-------------AGCCCG-GGGACAUUAAUCAUUAGGC-- ((((((((--.....-..))))(((..((((((.((.....)).)))))).((((((....))))))--.))-)-------------)))).(-....)..............-- ( -24.10) >DroSim_CAF1 59601 96 - 1 AAAUGGCACACUACA-AAUGUCGCCGUCAAUUAAUUUCCAUAGAUAAUUGCCUGUCGAUAACGACAG--CAG-C-------------AGGC-G-GGGACAUUAAUCAAAAGGCCG ....(((........-((((((.(((((........((....))....(((((((((....))))))--..)-)-------------))))-)-).)))))).........))). ( -30.63) >DroWil_CAF1 88817 104 - 1 -----GGAAACUAGAGAAUGUCGCCAACAAUUAAUUUCCAUAGAUAAUUGCCUGUCGAUAACAACAAAGCGGGCAAAAGAAAUGCCUAGC--C-GGAACAUUAAUCAAAUGU--- -----((..((........))..))....((((((((((...((((......))))............(((((((.......))))).))--.-)))).)))))).......--- ( -18.70) >DroAna_CAF1 62840 98 - 1 GGCUGGCACACUACA-AAUGUCGCCAACAAUUAAUUUCCAUAGAUAAUUGCCUGUCGAUAACGACAA--CGG-CAAC----------AGCC-GGGGGACAUUAAUCAAAUGUC-- ((((((((((.....-..))).)))......................((((((((((....))))).--.))-))).----------))))-....((((((.....))))))-- ( -26.50) >DroPer_CAF1 61011 95 - 1 GGCUGGCACACAACA-AAUGUCGCCAACAAUUAAUUUCCAUAGAUAAUUGCCUGUCGAUAACGACAG--CGG-C-------------AGCCCG-GGGACAUUAAUCAUUAGGC-- ((((((((.......-..))))(((..((((((.((.....)).)))))).((((((....))))))--.))-)-------------)))).(-....)..............-- ( -23.60) >consensus GGCUGGCACACUACA_AAUGUCGCCAACAAUUAAUUUCCAUAGAUAAUUGCCUGUCGAUAACGACAG__CGG_C_____________AGCC_G_GGGACAUUAAUCAAAAGGC__ ................((((((.((..((((((.((.....)).)))))).((((((....))))))...........................))))))))............. (-13.65 = -14.32 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:36 2006