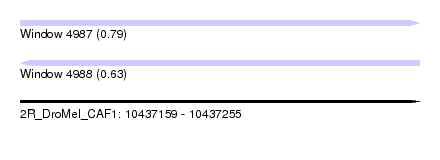
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,437,159 – 10,437,255 |
| Length | 96 |
| Max. P | 0.791191 |

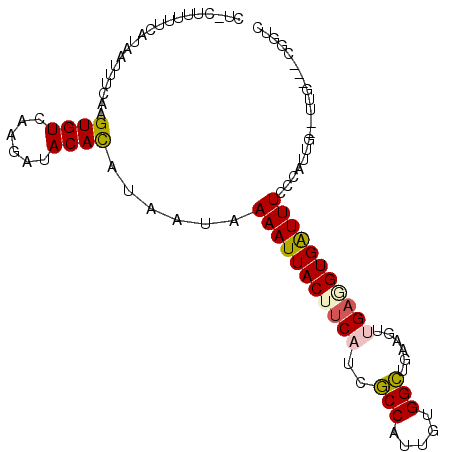
| Location | 10,437,159 – 10,437,255 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 76.68 |
| Mean single sequence MFE | -17.15 |
| Consensus MFE | -11.86 |
| Energy contribution | -12.75 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
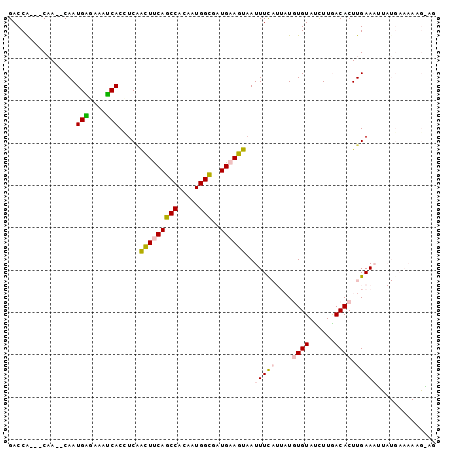
>2R_DroMel_CAF1 10437159 96 + 20766785 GACCA---CAA--CAAUGAGAAAUCACUACCAUUUCAGCCACAAUGGCGAUGAAGUAAUUUUAUUAUGUGUAUCUUGACACUUGAAAUUAUGAAAAAGAAG .....---...--...(((....)))......((((.(((.....)))......(((((((((....((((......)))).)))))))))))))...... ( -17.40) >DroPse_CAF1 37487 80 + 1 GACCGCCGCCA--CAAUGGGAAACCACAUCAACUCCAUCCACAAUGGAAAUGAAGUAAUUUC----UAUGUAUUUUAACACUUGAA--------------- ...........--...(((....)))..((((...........((((((((......)))))----)))............)))).--------------- ( -12.80) >DroSim_CAF1 39691 96 + 1 GACCA---CAA--CAAUGAGAAAUCACCUCGGCUUCAGCCACAAUGGCGAUGAAGUAAUUUUAUUAUGUGUACCUUGACACUUGAAAUCAUGAAAAAGAAG .....---...--...(((....)))..((((((((((((.....)))..)))))).((((((....((((......)))).))))))..)))........ ( -16.90) >DroEre_CAF1 38858 96 + 1 GACCG---CAA--CAAUGGGAAAUCACCUCAACUUCAGCCACAAUGGCGAUGAAGUAAUUUUAUUAUGUGUAUCUUGACACUUGAAACUAUGAAAAAGGAG ..((.---...--..((((............(((((((((.....)))..)))))).......(((.((((......)))).)))..))))......)).. ( -18.40) >DroYak_CAF1 41117 98 + 1 GACCA---CAACACAAUGAGAAAUCACCUCAACUUCAGCCACAAUGGCGUUGAAGUAAUUUCAUUAUGUGUAUCUUGACACUUGAAAUUAUGAAAAAGGAG ..((.---((((....((((.......))))......(((.....)))))))..(((((((((....((((......)))).)))))))))......)).. ( -24.60) >DroPer_CAF1 43704 80 + 1 GACCGCCGCCA--CAAUGGGAAACCACAUCAACUCCAUCCACAAUGGAAAUGAAGUAAUUUC----UAUGUAUUUUAACACUUGAA--------------- ...........--...(((....)))..((((...........((((((((......)))))----)))............)))).--------------- ( -12.80) >consensus GACCA___CAA__CAAUGAGAAAUCACCUCAACUUCAGCCACAAUGGCGAUGAAGUAAUUUCAUUAUGUGUAUCUUGACACUUGAAAUUAUGAAAAAG_AG ................(((....))).....(((((((((.....)))..))))))..(((((....((((......)))).))))).............. (-11.86 = -12.75 + 0.89)

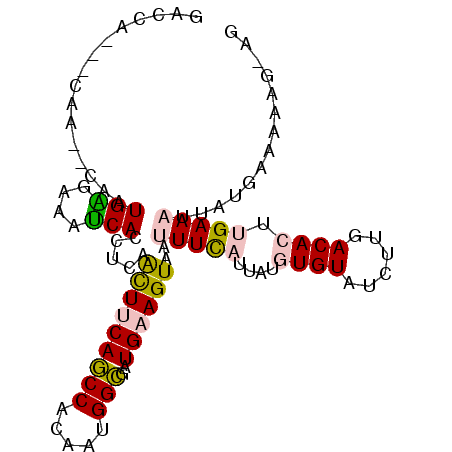
| Location | 10,437,159 – 10,437,255 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 76.68 |
| Mean single sequence MFE | -21.42 |
| Consensus MFE | -12.92 |
| Energy contribution | -12.78 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10437159 96 - 20766785 CUUCUUUUUCAUAAUUUCAAGUGUCAAGAUACACAUAAUAAAAUUACUUCAUCGCCAUUGUGGCUGAAAUGGUAGUGAUUUCUCAUUG--UUG---UGGUC ....................((((....)))).((((((((.......((((..(((((........)))))..)))).......)))--)))---))... ( -19.64) >DroPse_CAF1 37487 80 - 1 ---------------UUCAAGUGUUAAAAUACAUA----GAAAUUACUUCAUUUCCAUUGUGGAUGGAGUUGAUGUGGUUUCCCAUUG--UGGCGGCGGUC ---------------......((((....((((..----((((((((.(((.(((((((...))))))).))).))))))))....))--))..))))... ( -19.50) >DroSim_CAF1 39691 96 - 1 CUUCUUUUUCAUGAUUUCAAGUGUCAAGGUACACAUAAUAAAAUUACUUCAUCGCCAUUGUGGCUGAAGCCGAGGUGAUUUCUCAUUG--UUG---UGGUC ...((((..(((........)))..))))....((((((((.....(((((..(((.....))))))))..((((.....)))).)))--)))---))... ( -19.90) >DroEre_CAF1 38858 96 - 1 CUCCUUUUUCAUAGUUUCAAGUGUCAAGAUACACAUAAUAAAAUUACUUCAUCGCCAUUGUGGCUGAAGUUGAGGUGAUUUCCCAUUG--UUG---CGGUC .............((..(((((((......))))......((((((((((((((((.....))).))...)))))))))))....)))--..)---).... ( -20.30) >DroYak_CAF1 41117 98 - 1 CUCCUUUUUCAUAAUUUCAAGUGUCAAGAUACACAUAAUGAAAUUACUUCAACGCCAUUGUGGCUGAAGUUGAGGUGAUUUCUCAUUGUGUUG---UGGUC ........(((((.......((((....))))((((((((((((((((((((((((.....)))....))))))))))))..)))))))))))---))).. ( -29.70) >DroPer_CAF1 43704 80 - 1 ---------------UUCAAGUGUUAAAAUACAUA----GAAAUUACUUCAUUUCCAUUGUGGAUGGAGUUGAUGUGGUUUCCCAUUG--UGGCGGCGGUC ---------------......((((....((((..----((((((((.(((.(((((((...))))))).))).))))))))....))--))..))))... ( -19.50) >consensus CU_CUUUUUCAUAAUUUCAAGUGUCAAGAUACACAUAAUAAAAUUACUUCAUCGCCAUUGUGGCUGAAGUUGAGGUGAUUUCCCAUUG__UUG___CGGUC ....................((((......))))......(((((((((((..(((.....)))......))))))))))).................... (-12.92 = -12.78 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:29 2006