| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,418,478 – 10,418,580 |
| Length | 102 |
| Max. P | 0.798661 |

| Location | 10,418,478 – 10,418,580 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 78.27 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -21.59 |
| Energy contribution | -21.82 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
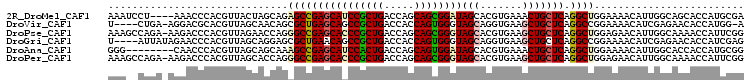
>2R_DroMel_CAF1 10418478 102 + 20766785 AAAUCCU----AAACCCACGUUACUAGCAGAGCCGAGCAUCCGCUGACCAGCAGCGGAUAGCACGUGAAACUGCUCAGGCUGGAAAACAUUGGCAGCACCAUGCGA .......----....(((.(((...(((((..(((.((((((((((.....)))))))).)).)).)...)))))..)))))).........(((......))).. ( -31.50) >DroVir_CAF1 30346 100 + 1 U----CUGA-AGGACGCACGUUAGCAACAGCGCUGAGCAGCCGCUGACCACCAGUGGGUAGCAGGUGAAGCUGCUCAGGCCGGAAAACAUCGAGAACACCAUGG-A (----(((.-.....((......))....((.(((((((((((((..(.(((....))).)..))))..)))))))))))))))....................-. ( -30.70) >DroPse_CAF1 20584 105 + 1 AAAGCCAGA-AAGACCCACGUUAGAACCAGGGCCGAGCACCCGCUGACCAGCAGCGGGUAGCACGUGAAGCUGCUCAGGCUGGAGAACAUUGGCAAAACCAUUCGG ...(((((.-..(((....)))....(((..(((((((((((((((.....))))))))(((.......))))))).))))))......)))))............ ( -37.10) >DroGri_CAF1 17694 102 + 1 U----AUUAUAGAACCCACGUUAGCAGGAGCGCUGAACAGCCGCUGACCACCAGUGGGUAGCAGGUGAAGCUGCUCAGGCCGGAAAACAUCGAGAACACCAUCGAG .----...............(..((..(((((((.....(((((((.(((....))).)))).)))..))).))))..))..)......((((........)))). ( -29.00) >DroAna_CAF1 26710 98 + 1 GGG--------CAACCCACGUUAGCAGCAAAGCCGAGCAUCCACUGACCAGCAGUGGAUAGCACGUGAAACUGCUCAGGCUGGAAAACAUUGGCACCACCAUGCGG ((.--------...))(((((..((......))...((((((((((.....)))))))).)))))))...((((...((.(((............)))))..)))) ( -31.40) >DroPer_CAF1 26670 105 + 1 AAAGCCAGA-AAGACCCACGUUAGCACCAGGGCCGAGCACCCGCUGACCAGCAGCGGGUAGCACGUGAAGCUGCUCAGGCUGGAGAACAUUGGCAAAACCAUUCGG ...(((((.-..(((....)))....(((..(((((((((((((((.....))))))))(((.......))))))).))))))......)))))............ ( -37.10) >consensus AAA__CUGA_AAAACCCACGUUAGCAGCAGAGCCGAGCACCCGCUGACCAGCAGCGGGUAGCACGUGAAGCUGCUCAGGCUGGAAAACAUUGGCAACACCAUGCGG ..............................((((((((((((((((.....))))))))(((.......))))))).))))......................... (-21.59 = -21.82 + 0.22)


| Location | 10,418,478 – 10,418,580 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 78.27 |
| Mean single sequence MFE | -36.97 |
| Consensus MFE | -25.24 |
| Energy contribution | -25.02 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
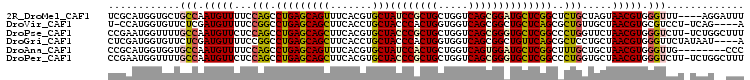
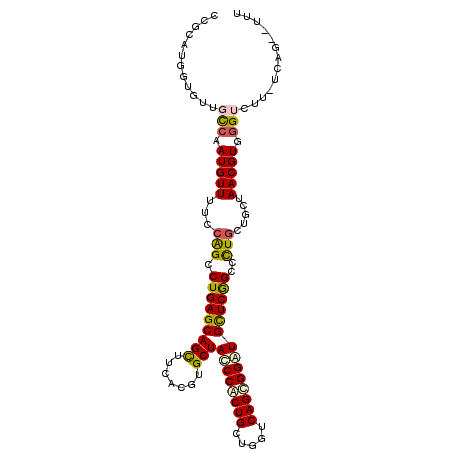
>2R_DroMel_CAF1 10418478 102 - 20766785 UCGCAUGGUGCUGCCAAUGUUUUCCAGCCUGAGCAGUUUCACGUGCUAUCCGCUGCUGGUCAGCGGAUGCUCGGCUCUGCUAGUAACGUGGGUUU----AGGAUUU ..((((((.....)).))))..((((((((.(((((.....((.((.((((((((.....)))))))))).))...)))))........))))).----.)))... ( -33.60) >DroVir_CAF1 30346 100 - 1 U-CCAUGGUGUUCUCGAUGUUUUCCGGCCUGAGCAGCUUCACCUGCUACCCACUGGUGGUCAGCGGCUGCUCAGCGCUGUUGCUAACGUGCGUCCU-UCAG----A .-.............(((((....((((((((((((((....((((((((....))))).))).)))))))))).))))..((....)))))))..-....----. ( -37.40) >DroPse_CAF1 20584 105 - 1 CCGAAUGGUUUUGCCAAUGUUCUCCAGCCUGAGCAGCUUCACGUGCUACCCGCUGCUGGUCAGCGGGUGCUCGGCCCUGGUUCUAACGUGGGUCUU-UCUGGCUUU .....(((.....))).........((((.(((((((.......)))((((((((.....))))))))))))(((((((.......)).)))))..-...)))).. ( -39.10) >DroGri_CAF1 17694 102 - 1 CUCGAUGGUGUUCUCGAUGUUUUCCGGCCUGAGCAGCUUCACCUGCUACCCACUGGUGGUCAGCGGCUGUUCAGCGCUCCUGCUAACGUGGGUUCUAUAAU----A .((((........))))........(((((((((((((....((((((((....))))).))).)))))))))).)))((..(....)..)).........----. ( -36.20) >DroAna_CAF1 26710 98 - 1 CCGCAUGGUGGUGCCAAUGUUUUCCAGCCUGAGCAGUUUCACGUGCUAUCCACUGCUGGUCAGUGGAUGCUCGGCUUUGCUGCUAACGUGGGUUG--------CCC ..((((((.....)).))))....((((((.((((((....((.((.((((((((.....)))))))))).)).....)))))).....))))))--------... ( -35.80) >DroPer_CAF1 26670 105 - 1 CCGAAUGGUUUUGCCAAUGUUCUCCAGCCUGAGCAGCUUCACGUGCUACCCGCUGCUGGUCAGCGGGUGCUCGGCCCUGGUGCUAACGUGGGUCUU-UCUGGCUUU .....(((.....))).........((((.(((((((.......)))((((((((.....))))))))))))(((((..((....))..)))))..-...)))).. ( -39.70) >consensus CCGCAUGGUGUUGCCAAUGUUUUCCAGCCUGAGCAGCUUCACGUGCUACCCACUGCUGGUCAGCGGAUGCUCGGCCCUGCUGCUAACGUGGGUCUU_UCAG__UUU ............(((.(((((...(((.(((((((((.......)))((((((((.....))))))))))))))..))).....))))).)))............. (-25.24 = -25.02 + -0.22)
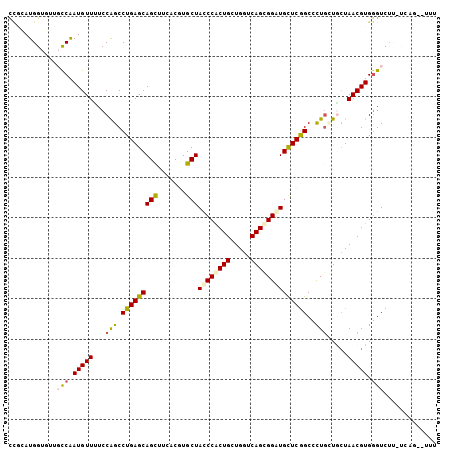
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:22 2006