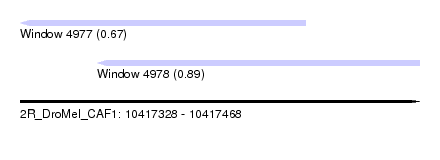
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,417,328 – 10,417,468 |
| Length | 140 |
| Max. P | 0.890969 |

| Location | 10,417,328 – 10,417,428 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 79.48 |
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -22.53 |
| Energy contribution | -22.25 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671411 |
| Prediction | RNA |
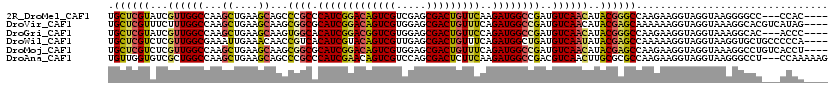
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10417328 100 - 20766785 GACAGUCGUCGAGCGACUGUUCAAGAUGGCCGAUGUCAACAUACGGGCCAAGAAGGUAGGUAAGGGGCC---CCACCG-CCCAUGGAUCUCGAGCCCCGUAUGU-- ((((((((.....)))))))).....((((....))))(((((((((.(..((.(((.......((((.---.....)-)))....)))))..).)))))))))-- ( -36.30) >DroPse_CAF1 19458 99 - 1 GACAGUCGUGGAGCGACUGUUCAAGAUGGCCGAUGUCAACAUACGCGCCAAGAAGGUAGGUAAGGGGCC---GCACAACCCCAGAGACCACAGAUCCAGA--CA-- ....(((.((((....((((......(((((((((....))).)).))))........(((..((((..---......))))....))))))).))))))--).-- ( -31.00) >DroEre_CAF1 19673 99 - 1 AACAGUCGUCGAGCGACUGUUCAAGAUGGCCGACGUCAACAUACGGGCCAAGAAGGUAGGUAAGGGGC----CCAACG-CCCAGUGAUCCCGAGCCCAGUAUGA-- ((((((((.....))))))))...((((.....))))..(((((((((......((..(((...((((----.....)-)))....)))))..)))).))))).-- ( -35.60) >DroYak_CAF1 21333 100 - 1 AACAGUCGUCGAGCGACUGUUCAAGAUGGCCGAUGUCAACAUACGGGCCAAGAAGGUAGGUAAGGGGCC---CCAACG-CCCACUGAUCCCGAGCCCAGUUUGA-- .(((((((.....)))))))(((((..(((((..((((.(.(((..(((.....)))..))).)((((.---.....)-)))..))))..)).)))...)))))-- ( -34.90) >DroMoj_CAF1 27529 103 - 1 GACAGUCGUGGAGCGACUGUUUCAGAUGGCCGAUGUCAACAUACGAGCCAAGAAGGUAGGUAAAGGCCUGUCACCUGG-UCGAUUGGCAUUGCAU-CACUC-AUAC ((((((((.....))))))))...((((..(((((((((....(((.(((.(..(..((((....))))..)..))))-))).))))))))))))-)....-.... ( -35.40) >DroPer_CAF1 25535 99 - 1 GACAGUCGUGGAGCGACUGUUCAAGAUGGCCGAUGUCAACAUACGCGCCAAGAAGGUAGGUAAGGGGCC---GCACAACCCCAGAGACCACAGAUCCUGA--CA-- .(((((((.....)))))))(((.(((((((((((....))).)).)))......((.(((..((((..---......))))....)))))..))).)))--..-- ( -32.00) >consensus GACAGUCGUCGAGCGACUGUUCAAGAUGGCCGAUGUCAACAUACGGGCCAAGAAGGUAGGUAAGGGGCC___CCACCG_CCCAGUGACCACGAACCCAGU__GA__ ((((((((.....)))))))).....(((((((((....))).)).))))....(((.(((...(((............)))....)))....))).......... (-22.53 = -22.25 + -0.28)

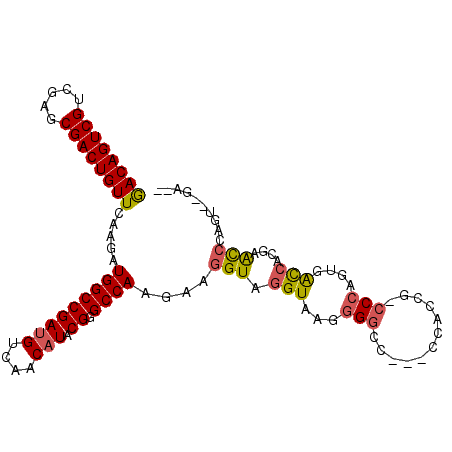
| Location | 10,417,355 – 10,417,468 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.44 |
| Mean single sequence MFE | -43.78 |
| Consensus MFE | -33.68 |
| Energy contribution | -33.80 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
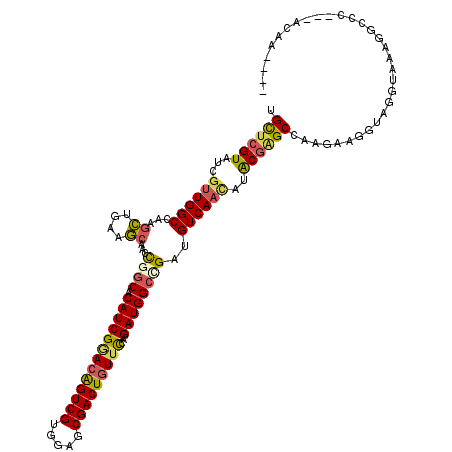
>2R_DroMel_CAF1 10417355 113 - 20766785 UGCUCGUAUCGUUGGCCAAGCUGAAGCAGCCCGCCCAUCGGACAGUCGUCGAGCGACUGUUCAAGAUGGCCGAUGUCAACAUACGGGCCAAGAAGGUAGGUAAGGGGCC---CCAC---- .((((((((.((((((...((((...)))).((.((((((((((((((.....)))))))))..))))).))..))))))))))))))......((..(((.....)))---))..---- ( -46.90) >DroVir_CAF1 29048 116 - 1 UGCUCGUUUCUUUGGCCAAGCUGAAGCAAGCGGCGCAUCGGACAGUCGUGGAGCGACUGUUUCAGAUGGCCGAUGUCAACAUACGAGCAAAAAAGGUAGGUAAAGGCACGUCAUAG---- (((((((....(((((...(((......)))(((.((((.((((((((.....))))))))...)))))))...)))))...)))))))...........................---- ( -40.90) >DroGri_CAF1 16570 113 - 1 UGCUCGUAUCGUUGGCCAAGCUGAAGCAAGUGGCACAUCGGACGGUCGUGGAGCGACUGUUCCAGAUGGCCGAUGUCAACAUACGGGCCAAGAAGGUAGGUAAAGGCAC---ACCC---- .((((((((.((((((((.((....))...))))(((((((.(.(((.((((((....))))))))).))))))))))))))))))))......(((..((....))..---))).---- ( -48.30) >DroWil_CAF1 37277 116 - 1 UGCUCGUCUCGUUGGCGAAAUUGAAACAACCGUCACAUCGUACAGUCGUUGAGCGACUGUUUCAGAUGGCUGAUGUCAAUAUACGAGCCAAAAAGGUAGGUAAGGUGCUGCCCCCA---- .((((((.(((....))).(((((..((.(((((.......(((((((.....)))))))....))))).))...)))))..))))))......(((((........)))))....---- ( -36.50) >DroMoj_CAF1 27556 116 - 1 UGCUCGUCUCGUUGGCCAAGCUGAAGCAAGCGGCGCAUCGGACAGUCGUGGAGCGACUGUUUCAGAUGGCCGAUGUCAACAUACGAGCCAAGAAGGUAGGUAAAGGCCUGUCACCU---- .((((((...((((((...(((......)))(((.((((.((((((((.....))))))))...)))))))...))))))..))))))......(..((((....))))..)....---- ( -48.90) >DroAna_CAF1 25608 117 - 1 UGUUGGUGUCGCUGGCCAAGCUGAAGCAGCCCGCCCAUCGAACAGUCGUCCAGCGACUCUUCAAGAUGGCCGACGUCAACUUGCGCGCCAAGAAGGUAGGUAAGGGCCU---CCAAAAAG (.((((((.(((((((...((((...)))).((.((((((((.(((((.....))))).)))..))))).))..))))....))))))))).).((.((((....))))---))...... ( -41.20) >consensus UGCUCGUAUCGUUGGCCAAGCUGAAGCAACCGGCACAUCGGACAGUCGUGGAGCGACUGUUCCAGAUGGCCGAUGUCAACAUACGAGCCAAGAAGGUAGGUAAAGGCCC___ACAA____ .((((((...((((((...((....))...((((.(((((((((((((.....)))))))))..))))))))..))))))..))))))................................ (-33.68 = -33.80 + 0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:20 2006