| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,415,703 – 10,415,823 |
| Length | 120 |
| Max. P | 0.960579 |

| Location | 10,415,703 – 10,415,823 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.50 |
| Mean single sequence MFE | -46.88 |
| Consensus MFE | -22.35 |
| Energy contribution | -24.72 |
| Covariance contribution | 2.37 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960579 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
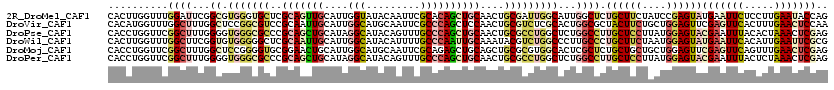
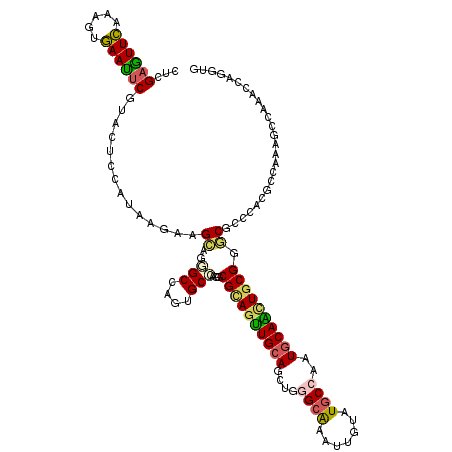
>2R_DroMel_CAF1 10415703 120 + 20766785 CACUUGGUUUGGAUUCGGCGUGGGUGCUCGCAGUUGCAUUGGUAUACAAUUCGCACAGCUGCAACUGCGAUUGGCAUUGGCUCUGCUUCUAUCCGAGUAUGAAUUCUCCUUGAAUACCAG .((((((..((((..(((.((.(((((((((((((((((((((.........)).))).)))))))))))...))))).)).)))..)))).))))))....((((.....))))..... ( -44.20) >DroVir_CAF1 27072 120 + 1 CACAUGGUUUGGCUUUGGCUCCGGCGUCCGCAAUUGCAUUGGCAUGCAAUUCGCCCAGCUCCAACUGCGUCUCGCACUGGCGCUACUUCUGCUGGAGUUCGAGUUCACUUUGAACUCCAA ..((.((.....)).))(((((((((..((.((((((((....))))))))))..)(((.(((..((((...)))).))).)))......))))))))..((((((.....))))))... ( -41.50) >DroPse_CAF1 18038 120 + 1 CACCUGGUUCGGCUUUGGGGUGGGCGCCCGCAGCUGCAUAGGCAUACAGUUUGCCCAGCUGCAACUGCGCCUGGCUCUGGCCUUGCUCCUUAUGGAGUACGAAUUUACACUAAACUCGAG .....(((((((((..(((.(((((((..(((((((....((((.......)))))))))))....))))))).))).)))).((((((....)))))).)))))............... ( -50.10) >DroWil_CAF1 35764 120 + 1 CACUUGGUUUGGCUUCGGUGUGGGGGCUCGCAAUUGCAUUGGCAUACAUUUUGCCCAAUUGCAAAUACGUCUGGCCCUUGCCCUGCUUCUAAUGGAGUAUGAAUUCACAUUGAAUUCGCG .((((.(((.(((...((.(..((((((((...(((((..((((.......))))....)))))...))...))))))..))).)))...))).))))..((((((.....))))))... ( -41.60) >DroMoj_CAF1 24972 120 + 1 CACCUGGUUCGGCUUUGGCUCCGGGGUGCGGAACUGCAUUGGCAUGCAAUUCGCAGAGCUGCAGCUGCGCGUGGCACUCGCUCUGCUGCUGCUGGAGUUCGAGUUCAGUUUGAACUCGAG ..(((((...(((....)))))))).((((((..(((((....))))).))))))(((((.((((.(((((.(((....))).))).)).)))).)))))((((((.....))))))... ( -53.00) >DroPer_CAF1 24115 120 + 1 CACCUGGUUCGGCUUUGGGGUGGGCGCCCGCAGCUGCAUAGGCAUACAGUUUGCCCAGCUGCAACUGCGCCUGGCUCUGGCCUUGCUCCUUAUGGAGUACGAAUUUACUCUAAACUCGAG ........(((..((((((((((((((..(((((((....((((.......)))))))))))....))))))(((....))).((((((....)))))).......))))))))..))). ( -50.90) >consensus CACCUGGUUCGGCUUUGGCGUGGGCGCCCGCAACUGCAUUGGCAUACAAUUCGCCCAGCUGCAACUGCGCCUGGCACUGGCCCUGCUUCUAAUGGAGUACGAAUUCACUUUGAACUCGAG ..........((((...((.(((((((..(((((((....(((.........))))))))))....)))))))))...)))).((((((....))))))(((((((.....))))))).. (-22.35 = -24.72 + 2.37)
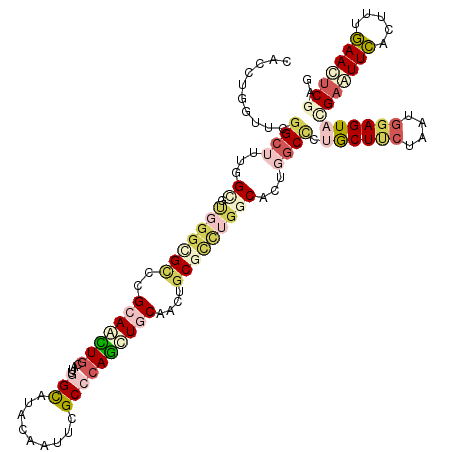
| Location | 10,415,703 – 10,415,823 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.50 |
| Mean single sequence MFE | -41.48 |
| Consensus MFE | -25.02 |
| Energy contribution | -24.50 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10415703 120 - 20766785 CUGGUAUUCAAGGAGAAUUCAUACUCGGAUAGAAGCAGAGCCAAUGCCAAUCGCAGUUGCAGCUGUGCGAAUUGUAUACCAAUGCAACUGCGAGCACCCACGCCGAAUCCAAACCAAGUG .((((.......(((........)))((((....(((.......)))...(((((((((((..(((((.....)))))....)))))))))))((......))...))))..)))).... ( -33.90) >DroVir_CAF1 27072 120 - 1 UUGGAGUUCAAAGUGAACUCGAACUCCAGCAGAAGUAGCGCCAGUGCGAGACGCAGUUGGAGCUGGGCGAAUUGCAUGCCAAUGCAAUUGCGGACGCCGGAGCCAAAGCCAAACCAUGUG .((((((((..((....)).))))))))((.....((((.((((((((...)))).)))).))))(((.((((((((....)))))))).(((...)))..)))...))........... ( -42.50) >DroPse_CAF1 18038 120 - 1 CUCGAGUUUAGUGUAAAUUCGUACUCCAUAAGGAGCAAGGCCAGAGCCAGGCGCAGUUGCAGCUGGGCAAACUGUAUGCCUAUGCAGCUGCGGGCGCCCACCCCAAAGCCGAACCAGGUG ..((((((((...))))))))..((((....))))...(((....))).((((((((((((..((((((.......)))))))))))))))(((.(....))))...))).......... ( -45.30) >DroWil_CAF1 35764 120 - 1 CGCGAAUUCAAUGUGAAUUCAUACUCCAUUAGAAGCAGGGCAAGGGCCAGACGUAUUUGCAAUUGGGCAAAAUGUAUGCCAAUGCAAUUGCGAGCCCCCACACCGAAGCCAAACCAAGUG (((((((((.....))))))..............((.((....((((....((((.(((((....((((.......))))..))))).)))).)))).....))...))........))) ( -31.90) >DroMoj_CAF1 24972 120 - 1 CUCGAGUUCAAACUGAACUCGAACUCCAGCAGCAGCAGAGCGAGUGCCACGCGCAGCUGCAGCUCUGCGAAUUGCAUGCCAAUGCAGUUCCGCACCCCGGAGCCAAAGCCGAACCAGGUG .((((((((.....)))))))).(((((((.(((((...(((.......)))...))))).))).((((((((((((....)))))))).))))....)))).....(((......))). ( -50.00) >DroPer_CAF1 24115 120 - 1 CUCGAGUUUAGAGUAAAUUCGUACUCCAUAAGGAGCAAGGCCAGAGCCAGGCGCAGUUGCAGCUGGGCAAACUGUAUGCCUAUGCAGCUGCGGGCGCCCACCCCAAAGCCGAACCAGGUG ..((((((((...))))))))..((((....))))...(((....))).((((((((((((..((((((.......)))))))))))))))(((.(....))))...))).......... ( -45.30) >consensus CUCGAGUUCAAAGUGAAUUCGUACUCCAUAAGAAGCAGGGCCAGUGCCAGGCGCAGUUGCAGCUGGGCAAAUUGUAUGCCAAUGCAACUGCGGGCGCCCACGCCAAAGCCAAACCAGGUG ...((((((.....))))))..............((..(((....)))...((((((((((....((((.......))))..)))))))))).))......................... (-25.02 = -24.50 + -0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:18 2006