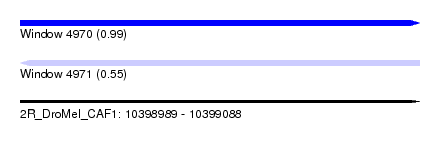
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,398,989 – 10,399,088 |
| Length | 99 |
| Max. P | 0.991146 |

| Location | 10,398,989 – 10,399,088 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 76.82 |
| Mean single sequence MFE | -20.30 |
| Consensus MFE | -13.04 |
| Energy contribution | -14.85 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
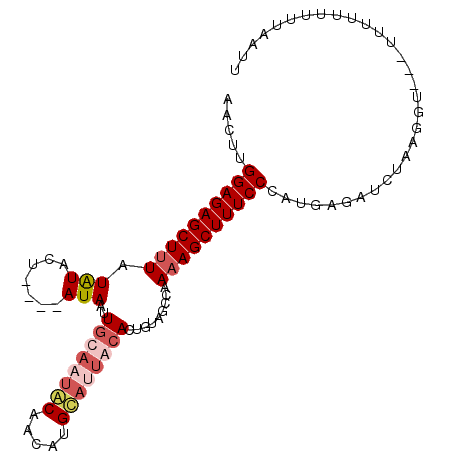
>2R_DroMel_CAF1 10398989 99 + 20766785 AACUUGGAGAGCUUUAUAUACUACAUAUAAUUGCAAUGCAACAAGCAUUACACUUCAUCCAAAAGCUUUCCCAUGAGAUCUAAGGU---UUUUUUUUUAAUU ..(((((((((((((.((((.....))))..((.(((((.....))))).)).........))))))))))...))).........---............. ( -17.60) >DroSim_CAF1 11373 95 + 1 AACUUGGAGAGCUUUAUGUACU----AUAAUUGCUAAGCAACAUGCAUUAGACGUUAUCCAAAAGCUUUCCCAUGAAAUCUAAGGU---UUUUUUUUUAAUU .....((((((((((.((....----(((((..((((((.....)).))))..))))).))))))))))))...(((((.....))---))).......... ( -18.60) >DroEre_CAF1 9967 94 + 1 UACCAGGAGAGCUAUGUAUACU----AUAUUUGCAUUACAAAAUGAAUUGCACUGUAGUCAAAAGCUUUCCUAUGAGAUCGAAGU----UUUUUUUGUAAUU ....(((((((((......(((----(((..((((.............)))).))))))....))))))))).......(((((.----...)))))..... ( -19.52) >DroYak_CAF1 11369 98 + 1 UACUUGGAGAGCUUUGUAUACU----AUAACUGCAUUACACUUUGUAUUGCACUGUAGCCAAAAGCUUUCCUAUGAGAAUUAAUUCUUUUUUUUUUCUAAUU .....((((((((((.....((----(((..((((.(((.....))).)))).)))))...))))))))))...(((((..((.....))..)))))..... ( -25.50) >consensus AACUUGGAGAGCUUUAUAUACU____AUAAUUGCAAUACAACAUGCAUUACACUGUAGCCAAAAGCUUUCCCAUGAGAUCUAAGGU___UUUUUUUUUAAUU .....((((((((((.(((.......)))..((((((((.....)))))))).........))))))))))............................... (-13.04 = -14.85 + 1.81)

| Location | 10,398,989 – 10,399,088 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 76.82 |
| Mean single sequence MFE | -17.91 |
| Consensus MFE | -12.06 |
| Energy contribution | -13.62 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
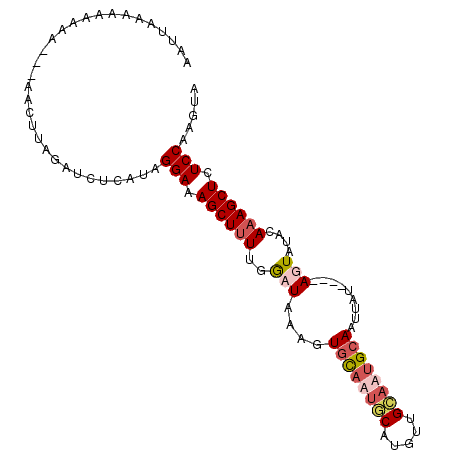
>2R_DroMel_CAF1 10398989 99 - 20766785 AAUUAAAAAAAAA---ACCUUAGAUCUCAUGGGAAAGCUUUUGGAUGAAGUGUAAUGCUUGUUGCAUUGCAAUUAUAUGUAGUAUAUAAAGCUCUCCAAGUU .............---...............(((.((((((...(((...((((((((.....))))))))...........)))..)))))).)))..... ( -19.74) >DroSim_CAF1 11373 95 - 1 AAUUAAAAAAAAA---ACCUUAGAUUUCAUGGGAAAGCUUUUGGAUAACGUCUAAUGCAUGUUGCUUAGCAAUUAU----AGUACAUAAAGCUCUCCAAGUU .............---...............(((.((((((((.((((...((((.((.....))))))...))))----....)).)))))).)))..... ( -15.30) >DroEre_CAF1 9967 94 - 1 AAUUACAAAAAAA----ACUUCGAUCUCAUAGGAAAGCUUUUGACUACAGUGCAAUUCAUUUUGUAAUGCAAAUAU----AGUAUACAUAGCUCUCCUGGUA .............----............(((((.((((....((((...((((...(.....)...))))....)----)))......)))).)))))... ( -16.60) >DroYak_CAF1 11369 98 - 1 AAUUAGAAAAAAAAAAGAAUUAAUUCUCAUAGGAAAGCUUUUGGCUACAGUGCAAUACAAAGUGUAAUGCAGUUAU----AGUAUACAAAGCUCUCCAAGUA ...............................(((.((((((..((((.((((((.(((.....))).)))).)).)----)))....)))))).)))..... ( -20.00) >consensus AAUUAAAAAAAAA___AACUUAGAUCUCAUAGGAAAGCUUUUGGAUAAAGUGCAAUGCAUGUUGCAAUGCAAUUAU____AGUAUACAAAGCUCUCCAAGUA ...............................(((.((((((..(((....((((((((.....)))))))).........)))....)))))).)))..... (-12.06 = -13.62 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:13 2006