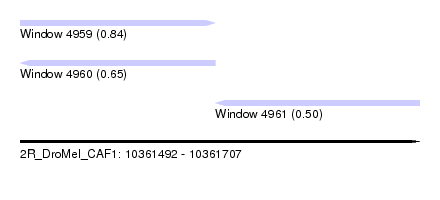
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,361,492 – 10,361,707 |
| Length | 215 |
| Max. P | 0.844228 |

| Location | 10,361,492 – 10,361,597 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.76 |
| Mean single sequence MFE | -46.09 |
| Consensus MFE | -24.24 |
| Energy contribution | -21.80 |
| Covariance contribution | -2.44 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
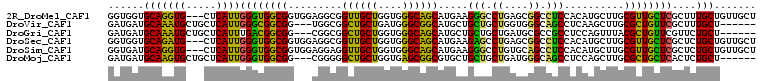
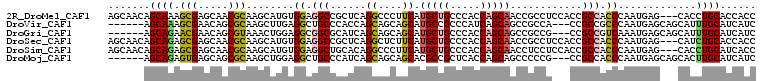
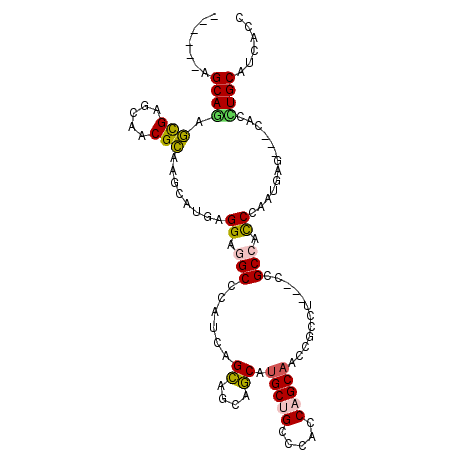
>2R_DroMel_CAF1 10361492 105 + 20766785 GGUGGUGCAGGUG---CUCAUUGGGUGGCGGUGGAGGCGGUUGCUGGUGGGCAGCAUGAAGGGCCUGAGCGGCCUCCACAUGCUUGCGUUGCUCGCUUUGCUGUUGCU (((((.((....)---)))))).((..((((..(((.(((..((....(.((((((((..(((((.....)))))...))))).))).).))))))))..))))..)) ( -42.20) >DroVir_CAF1 96415 99 + 1 GAUGAUGCAAAUGCUGCUCAUUGGGCGGCGG---UGGCGGCUGCUGAUGGGCGGCAUGCUGCUGCUGGUGGGCAGCCUCAAGCUUGCGCUGUUCGCUUUGCU------ ..(((.((...(((((((.....))))))).---..))(((((((.((.((((((.....)))))).)).))))))))))(((..(((.....)))...)))------ ( -49.00) >DroGri_CAF1 104225 99 + 1 GAUGAUGCAAAUGCUGCUCAUUUGACGGCGG---CGGCGGCUGCUGGUGGGCAGCAUGCUGCUGCUGAUGCGCCGCCUCCAGUUUACGCUGUUCGUUCUGCU------ (((((.(((.....)))))))).((((((((---.((((((.((..((.((((((.....)))))).)))))))))).)).(....))))))).........------ ( -41.90) >DroSec_CAF1 80341 105 + 1 GGUGGUGCAGAUG---CUCAUUGGGUGGCGGUGGAGGCGGUUGCUGGUGGGCAGCAUGAAGAGCCUGAGCGGCCUCCACAUGCUUGCGUUGCUCGCUCUGCUGUUGCU (((..((((((.(---(.....(((..(((((((((((.((((((....)))))).......((....)).)))))))).......)))..)))))))))).)..))) ( -42.11) >DroSim_CAF1 76369 105 + 1 GGUGAUGCAGGUG---CUCAUUGGGUGGCGGUGGAGGAGGUUGCUGGUGGGCAGCAUGAAGGGCCUGUGCAGCCUCCACAUGCUUGCGUUGCUCGCUCUGCUGUUGCU (((((.((....)---)))))).((..((((..(((((((((((..(..(((..(.....).)))..))))))))))........(((.....)))))..))))..)) ( -45.10) >DroMoj_CAF1 110020 99 + 1 GAUGAUGCAAGUGCUGCUCAUUGGGUGGCGG---CGGGGGCUGCUGGUGAGCGGCGUGCUGCUGCUGAUGGGCAGCCUCCAGCUUGCGCUGCUCACUCUGCU------ (((((.(((.....))))))))(((((((((---(((((((((((.((.((((((.....)))))).)).)))))))))).(....)))))).)))))....------ ( -56.20) >consensus GAUGAUGCAAAUG___CUCAUUGGGUGGCGG___AGGCGGCUGCUGGUGGGCAGCAUGAAGCGCCUGAGCGGCCGCCACAAGCUUGCGCUGCUCGCUCUGCU______ ......(((((((.....))))((((((((.........((((((....)))))).....(((.((....)).)))..........))))))))....)))....... (-24.24 = -21.80 + -2.44)
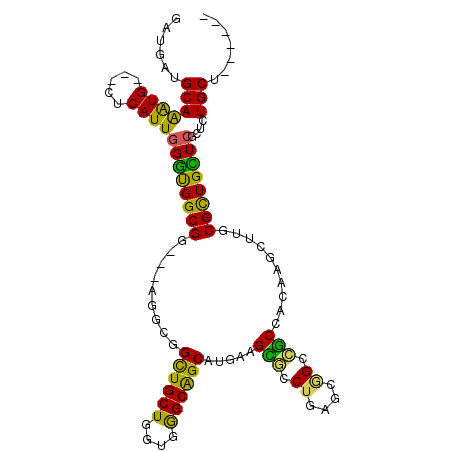
| Location | 10,361,492 – 10,361,597 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 75.76 |
| Mean single sequence MFE | -33.15 |
| Consensus MFE | -15.96 |
| Energy contribution | -15.63 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10361492 105 - 20766785 AGCAACAGCAAAGCGAGCAACGCAAGCAUGUGGAGGCCGCUCAGGCCCUUCAUGCUGCCCACCAGCAACCGCCUCCACCGCCACCCAAUGAG---CACCUGCACCACC .(((...((...(((.....)))..((..((((((((.((....))......(((((.....)))))...)))))))).))..........)---)...)))...... ( -32.20) >DroVir_CAF1 96415 99 - 1 ------AGCAAAGCGAACAGCGCAAGCUUGAGGCUGCCCACCAGCAGCAGCAUGCCGCCCAUCAGCAGCCGCCA---CCGCCGCCCAAUGAGCAGCAUUUGCAUCAUC ------.(((((((....(((....)))...((((((......((.((.....)).))......))))))))..---..((.((.......)).)).)))))...... ( -31.60) >DroGri_CAF1 104225 99 - 1 ------AGCAGAACGAACAGCGUAAACUGGAGGCGGCGCAUCAGCAGCAGCAUGCUGCCCACCAGCAGCCGCCG---CCGCCGUCAAAUGAGCAGCAUUUGCAUCAUC ------.((((((((.....)))....((..((((((((....)).((.((.(((((.....))))))).)).)---)))))..))...........)))))...... ( -34.90) >DroSec_CAF1 80341 105 - 1 AGCAACAGCAGAGCGAGCAACGCAAGCAUGUGGAGGCCGCUCAGGCUCUUCAUGCUGCCCACCAGCAACCGCCUCCACCGCCACCCAAUGAG---CAUCUGCACCACC .......((((((((.....)))..((..((((((((.((....))......(((((.....)))))...)))))))).))...........---..)))))...... ( -35.10) >DroSim_CAF1 76369 105 - 1 AGCAACAGCAGAGCGAGCAACGCAAGCAUGUGGAGGCUGCACAGGCCCUUCAUGCUGCCCACCAGCAACCUCCUCCACCGCCACCCAAUGAG---CACCUGCAUCACC .......((((.(((.....)))..((..(((((((..((....))......(((((.....)))))....))))))).))...........---...))))...... ( -28.40) >DroMoj_CAF1 110020 99 - 1 ------AGCAGAGUGAGCAGCGCAAGCUGGAGGCUGCCCAUCAGCAGCAGCACGCCGCUCACCAGCAGCCCCCG---CCGCCACCCAAUGAGCAGCACUUGCAUCAUC ------.(((.((((.(((((....)))((.((((((.....(((.((.....)).))).....)))))).)))---).((((.....)).))..)))))))...... ( -36.70) >consensus ______AGCAGAGCGAGCAACGCAAGCAUGAGGAGGCCCAUCAGCAGCAGCAUGCUGCCCACCAGCAACCGCCU___CCGCCACCCAAUGAG___CACCUGCAUCACC .......((((.(((.....)))........((.(((......((....)).(((((.....)))))............))).)).............))))...... (-15.96 = -15.63 + -0.33)
| Location | 10,361,597 – 10,361,707 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 81.27 |
| Mean single sequence MFE | -38.58 |
| Consensus MFE | -22.21 |
| Energy contribution | -24.05 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
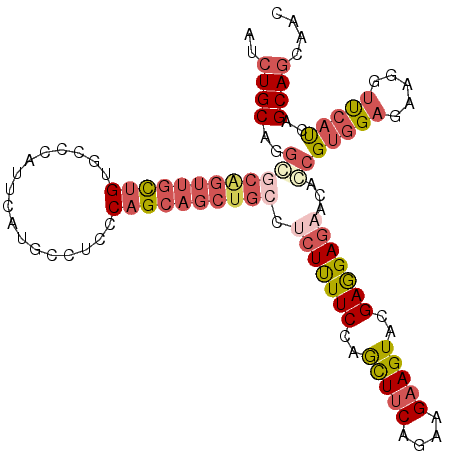
>2R_DroMel_CAF1 10361597 110 - 20766785 AUCUGCAGGCGCAGUUGCUGUGCCCAUUCAUACCUCCCAGCAGCUGCCUCUUUUCCAACUUCAGAAGAAGUACGAGGAGAACAGCGUGGAGAAGGUUCAUCAGCAGCAAC ..((((.(((((((...))))))).......((((((((...((((..(((((((..(((((....)))))..))))))).)))).))).).))))......)))).... ( -39.20) >DroSec_CAF1 80446 110 - 1 AUCUGCAGGCGCAGUUGUUGUGCCCAUUCAUGCCUCCCAGCAGCUGCCUCUUUUCCAGCUUCAGAAGAAGUACGAGGAGAACAGCGUGGAGAAGGUUCAUCAGCAGCAAC ..((((.(((((((...))))))).......((((((((...((((..(((((((..(((((....)))))..))))))).)))).))).).))))......)))).... ( -37.80) >DroSim_CAF1 76474 110 - 1 AUCUGCAGGCGCAGUUGCUGUGCCCAUUCAUGCCUCCCAGCAGCUGCCUCUUUUCCAACUUCAGAAGAAGUACGAAGAGACCAGCGUGGAGAAGGUUCAUCAGCAGCAAC ..((((.(((((((...))))))).......((((((((...((((..(((((((..(((((....)))))..))))))).)))).))).).))))......)))).... ( -39.10) >DroEre_CAF1 78107 110 - 1 GUCUGCAGGGGCAGUUGCUGUGCCCAUCCAUGCCUCCCAGCAGCUGCCUCUUUUCCAGCUUCAGAAGAAGUACGAGGAGAACACCGUGGAGAAGGUUCAUCAGCAGCAAC (.((((..((((((((((((.((........))....))))))))))))(((((((((((((....)))))....((......)).))))))))........)))))... ( -42.50) >DroAna_CAF1 80440 106 - 1 AGGUGCUGGAUCCGUUUCUGUUCCUCUACACGGUUCUCAGCAGCUACCGAUCUUCCAGUUGCAAAAGAAGUACGAGGAGAGUGCCGUGGACAAAGUCCACCAGCAG---- .((..(((((......))).(((((((((....((((..((((((...........))))))...))))))).))))))))..))((((((...))))))......---- ( -32.20) >DroPer_CAF1 85985 106 - 1 -UCUGCCGCCUCG---GCCGUGUCCCUACACGUCCCCCAGCAGCUGCCGCUCUUCCAGCUUCAGAAGAAGUACGAGGAGAGCGCCGCCGAGAAGGCUCACCAGCAGCAGC -.((((.((((((---(((((((....))))).............(.(((((((((.(((((....)))))..).)))))))).)))))))..((....)).)).)))). ( -40.70) >consensus AUCUGCAGGCGCAGUUGCUGUGCCCAUUCAUGCCUCCCAGCAGCUGCCUCUUUUCCAGCUUCAGAAGAAGUACGAGGAGAACACCGUGGAGAAGGUUCAUCAGCAGCAAC ..((((..((((((((((((.................)))))))))).(((((((..(((((....)))))..)))))))...))(((((.....)))))..)))).... (-22.21 = -24.05 + 1.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:03 2006