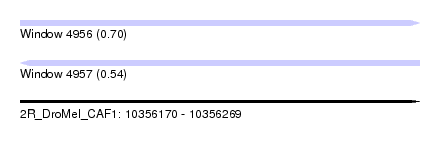
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,356,170 – 10,356,269 |
| Length | 99 |
| Max. P | 0.701789 |

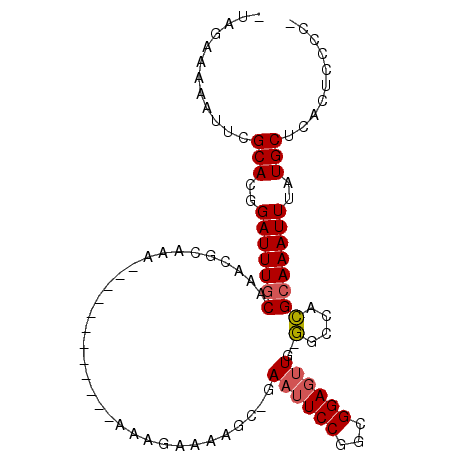
| Location | 10,356,170 – 10,356,269 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.86 |
| Mean single sequence MFE | -23.92 |
| Consensus MFE | -16.95 |
| Energy contribution | -17.90 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10356170 99 + 20766785 GGGGGAGUGAGCAUAAAUUUGCGUGGCC-CAACUCCACCGGAAUUC-GCUUUUCUUUUUUUUUCUUUUUUUUGCGUUUGCAAAUCCGUGCGAAUUUUCCUA- (((..(((..((((..((((((((((..-.....)))).......(-((.......................)))...))))))..))))..)))..))).- ( -25.60) >DroSec_CAF1 75038 87 + 1 GGGGGAGUGAGCAUAAAUUUGCGUGGCC-CAACUCCGCCGGAAUUC-GCUUUCCAUU------------UUUGCGUUUGCAAAUCCGUGCGAAUUUUCCUA- (((..(((..((((..(((((((.(((.-.......)))((((...-...))))...------------........)))))))..))))..)))..))).- ( -29.20) >DroSim_CAF1 71047 87 + 1 UGGGGGGUGAGCAUAAAUUUGCGUGGCC-CAACUCCGCCGGAAUUC-GCUUUCCAUU------------UUUGCGUUUGCAAAUCCGUGCGAAUUUUCCUA- .((..(((..((((..(((((((.(((.-.......)))((((...-...))))...------------........)))))))..))))..)))..))..- ( -28.00) >DroEre_CAF1 70812 86 + 1 -GGGGAGUGAGCAUAAAUUUGCGUAGCC-CAACUCCUCAGGAUUUC-GCUUUUCUUU------------UUCACGUUUGCAAAUCCGUGCGAAUUUUUCUG- -(((((((..(((......)))......-..))))))).((((((.-((........------------.........))))))))...............- ( -19.63) >DroYak_CAF1 72049 87 + 1 -GGGGAGUGAGCAUAAAUUUGCGUAGCC-CAACUCCUCCGGAAUUCCUUUUUUCUUU------------UUUACGCUUGCAAAUCCGUGCGAAUUUUUCUA- -(..((((..((((..(((((((.(((.-..........((((........))))..------------.....))))))))))..))))..))))..)..- ( -20.47) >DroPer_CAF1 80740 86 + 1 UGGGGAGUGAGCAUAAAUUUCCAUGGCUUCAGCUCCGGCGGAAUUU-CUCCUCCUUA------------UUUUCG---GCAAAUCCGUGCUAAAUUUUGUAU (((((((.(((...((((((((..(((....)))...).)))))))-))))))))))------------.....(---(((......))))........... ( -20.60) >consensus _GGGGAGUGAGCAUAAAUUUGCGUGGCC_CAACUCCGCCGGAAUUC_GCUUUCCUUU____________UUUGCGUUUGCAAAUCCGUGCGAAUUUUCCUA_ .(((((((..((((..((((((((((........)))).((((.......))))........................))))))..))))..)))))))... (-16.95 = -17.90 + 0.95)

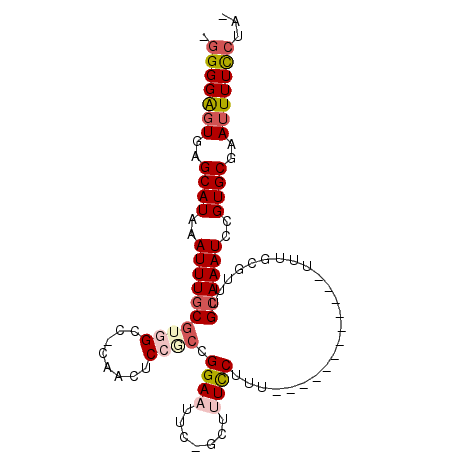
| Location | 10,356,170 – 10,356,269 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 80.86 |
| Mean single sequence MFE | -21.75 |
| Consensus MFE | -13.76 |
| Energy contribution | -13.98 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10356170 99 - 20766785 -UAGGAAAAUUCGCACGGAUUUGCAAACGCAAAAAAAAGAAAAAAAAAGAAAAGC-GAAUUCCGGUGGAGUUG-GGCCACGCAAAUUUAUGCUCACUCCCCC -..(((..((((((.....(((((....)))))....................))-)))))))((.(((((.(-(((.............))))))))))). ( -23.23) >DroSec_CAF1 75038 87 - 1 -UAGGAAAAUUCGCACGGAUUUGCAAACGCAAA------------AAUGGAAAGC-GAAUUCCGGCGGAGUUG-GGCCACGCAAAUUUAUGCUCACUCCCCC -..(((..((((((.((..(((((....)))))------------..))....))-)))))))((.(((((.(-(((.............))))))))))). ( -28.12) >DroSim_CAF1 71047 87 - 1 -UAGGAAAAUUCGCACGGAUUUGCAAACGCAAA------------AAUGGAAAGC-GAAUUCCGGCGGAGUUG-GGCCACGCAAAUUUAUGCUCACCCCCCA -..(((..((((((.((..(((((....)))))------------..))....))-)))))))((.((.(.((-(((.............))))).))))). ( -24.52) >DroEre_CAF1 70812 86 - 1 -CAGAAAAAUUCGCACGGAUUUGCAAACGUGAA------------AAAGAAAAGC-GAAAUCCUGAGGAGUUG-GGCUACGCAAAUUUAUGCUCACUCCCC- -............((.((((((((.....(...------------.)......))-.)))))))).(((((.(-(((.............)))))))))..- ( -18.12) >DroYak_CAF1 72049 87 - 1 -UAGAAAAAUUCGCACGGAUUUGCAAGCGUAAA------------AAAGAAAAAAGGAAUUCCGGAGGAGUUG-GGCUACGCAAAUUUAUGCUCACUCCCC- -...........(((..(((((((.(((.(((.------------..........((....)).......)))-.)))..)))))))..))).........- ( -18.57) >DroPer_CAF1 80740 86 - 1 AUACAAAAUUUAGCACGGAUUUGC---CGAAAA------------UAAGGAGGAG-AAAUUCCGCCGGAGCUGAAGCCAUGGAAAUUUAUGCUCACUCCCCA ...............(((.....)---))....------------...(((((((-........(((..((....))..))).........))).))))... ( -17.93) >consensus _UAGAAAAAUUCGCACGGAUUUGCAAACGCAAA____________AAAGAAAAGC_GAAUUCCGGCGGAGUUG_GGCCACGCAAAUUUAUGCUCACUCCCC_ ............(((..(((((((.................................((((((...))))))..(....))))))))..))).......... (-13.76 = -13.98 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:59 2006