
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,346,893 – 10,346,987 |
| Length | 94 |
| Max. P | 0.595582 |

| Location | 10,346,893 – 10,346,987 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.80 |
| Mean single sequence MFE | -21.87 |
| Consensus MFE | -15.79 |
| Energy contribution | -15.85 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10346893 94 - 20766785 UAGG-CGUGCAUUACUAAUUAUUGUAAUGAAAUUAUUUAUGAUAAUUGUCAGAGUCU--------CAGACACUGCACAUUGUUGAUCAUUCGGCGUGUGCACA----- ....-.((((((.((.((((((..(((.........)))..))))))(((((.(((.--------..))).))).))...(((((....))))))))))))).----- ( -23.40) >DroPse_CAF1 66312 95 - 1 UAAGUUUUGCAUUACUAAUUAUUGUAAUGGAAUUAUUUAUGAUCAUUAUCCGAUUGU--------CAGACACUGCACAUUGUUGUCCAUUCGGCGUGUGCACC----- (((((.((.((((((........)))))).))..)))))((((.(((....))).))--------)).....(((((((.((((......)))))))))))..----- ( -20.90) >DroEre_CAF1 61649 94 - 1 UAGG-CGUGCAUUACUAAUUAUUGUAAUGAAAUUAUUUAUGAUAAUUGGCAAAGUUU--------CAGACACUGCACAUUGUUGGGCAUUCGGCGUGUGCACA----- ....-.((((....((((((((..(((.........)))..))))))))........--------...((((.((....((.....))....)))))))))).----- ( -22.10) >DroMoj_CAF1 87808 105 - 1 UAGG-UAUGCAUUACUAAUUAUUGUAAUGGAAUUAUUUAUGAUUAUUGGCAAAGCCAACGGCGAUUUGACA-UGCACAUUGUUGAUCA-ACAGCUCGUGUAUUUAGCA ((((-((((((((((........)))))...........((((..(((((...)))))(((((((.((...-..)).)))))))))))-.......)))))))))... ( -20.60) >DroAna_CAF1 65853 94 - 1 UAGA-CGUGCAUUACUAAUCAUUGUAAUGGAAUUAUUUAUGAUAAUUGUCAAAGUCG--------CAGACACUGCACAUUGUUGGGCACUCGGCGUGUGCACA----- ..((-(...((((((........)))))).(((((((...))))))))))...(((.--------..)))..(((((((.(((((....))))))))))))..----- ( -23.30) >DroPer_CAF1 66668 95 - 1 UAAGUUUUGCAUUACUAAUUAUUGUAAUGGAAUUAUUUAUGAUCAUUAUCCGAUUGU--------CAGACACUGCACAUUGUUGUCCAUUCGGCGUGUGCACC----- (((((.((.((((((........)))))).))..)))))((((.(((....))).))--------)).....(((((((.((((......)))))))))))..----- ( -20.90) >consensus UAGG_CGUGCAUUACUAAUUAUUGUAAUGGAAUUAUUUAUGAUAAUUGUCAAAGUCU________CAGACACUGCACAUUGUUGACCAUUCGGCGUGUGCACA_____ .........((((((........))))))...........................................(((((((.((((......)))))))))))....... (-15.79 = -15.85 + 0.06)

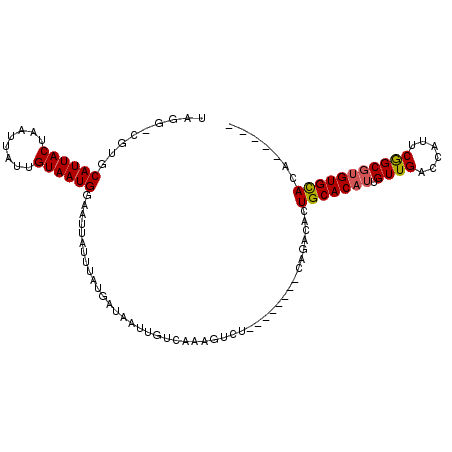
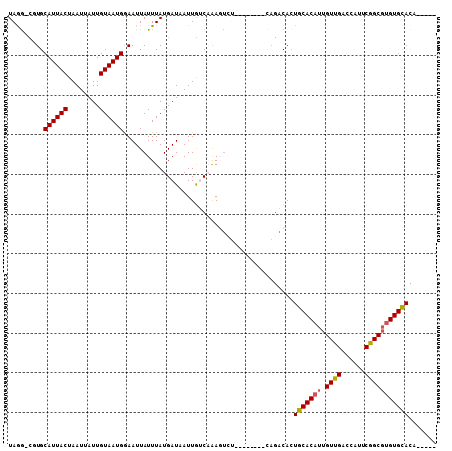
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:56 2006