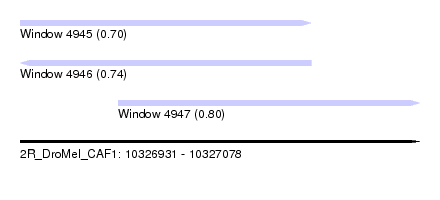
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,326,931 – 10,327,078 |
| Length | 147 |
| Max. P | 0.795015 |

| Location | 10,326,931 – 10,327,038 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 89.56 |
| Mean single sequence MFE | -26.88 |
| Consensus MFE | -19.08 |
| Energy contribution | -22.07 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
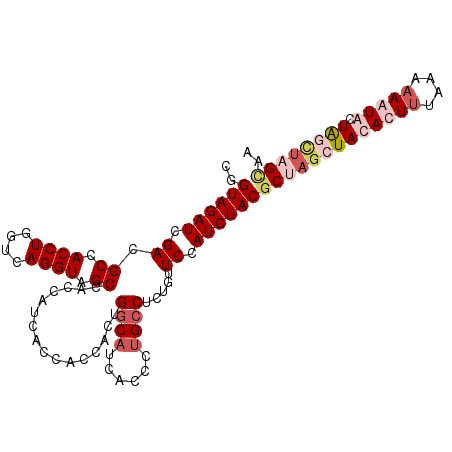
>2R_DroMel_CAF1 10326931 107 + 20766785 CGGUAGAUCGACGCCACCUGGUCAGGUAGCCACCAUCACCACCACUGGCAUCACCCUGCCUCUGUUCCAUCUACGCUAGAUACACUUUAAAAAAUGUUGGUAAGUAA .(((.(((....((.((((....)))).))....))))))((((..((((......))))........(((((...)))))................))))...... ( -21.60) >DroSec_CAF1 41392 107 + 1 CGGUAGAUCGACGCCACCUGGUCAGGUAGCCACCAUCACCACCAUUGGCAUCACCCUGCCUCGGUUCCAUCUACGCUAGCUACACUUUAAAAAGUACUAGCUUGCUA ..((((((.((.(((...((((..(((....)))...)))).....((((......))))..))))).))))))((.(((((.((((....))))..))))).)).. ( -30.80) >DroSim_CAF1 41584 107 + 1 CGGUAGAUCGACGCCACCUGGUCAGGUAGCCACCAUCACCACCACUGGCAUCACCCUGCCUCUGUUCCAUCUACGCUAGCUACACUUUAAAAAGUACUAGCUUGCAA ..((((((.((.((.((((....)))).))............((..((((......))))..)).)).))))))((.(((((.((((....))))..))))).)).. ( -29.50) >DroEre_CAF1 41382 107 + 1 CGGUAGAUCGACGCCACCUGGUCAGGUAGCCACCAUCACCACCACCGCCAUCAGCCCGCCUCUGCUCCAUCUACCCUAGCUACACUCUAAAAAAUGCUGUGUAGUAA .(((((((.((.((....((((..(((....)))...)))).....((.....))........)))).)))))))...(((((((.............))))))).. ( -25.62) >consensus CGGUAGAUCGACGCCACCUGGUCAGGUAGCCACCAUCACCACCACUGGCAUCACCCUGCCUCUGUUCCAUCUACGCUAGCUACACUUUAAAAAAUACUAGCUAGCAA ..((((((.((.((.((((....)))).))................((((......)))).....)).))))))(((((((((((((....))))).)))))))).. (-19.08 = -22.07 + 3.00)

| Location | 10,326,931 – 10,327,038 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 89.56 |
| Mean single sequence MFE | -37.03 |
| Consensus MFE | -29.85 |
| Energy contribution | -31.97 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.735838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10326931 107 - 20766785 UUACUUACCAACAUUUUUUAAAGUGUAUCUAGCGUAGAUGGAACAGAGGCAGGGUGAUGCCAGUGGUGGUGAUGGUGGCUACCUGACCAGGUGGCGUCGAUCUACCG ......((((............((.((((((...))))))..))...((((......))))..))))((((((.(..(((((((....)))))))..).))).))). ( -32.40) >DroSec_CAF1 41392 107 - 1 UAGCAAGCUAGUACUUUUUAAAGUGUAGCUAGCGUAGAUGGAACCGAGGCAGGGUGAUGCCAAUGGUGGUGAUGGUGGCUACCUGACCAGGUGGCGUCGAUCUACCG ..((.(((((.(((((....)))))))))).))((((((.((((((.((((......))))..))))..........(((((((....))))))).)).)))))).. ( -41.20) >DroSim_CAF1 41584 107 - 1 UUGCAAGCUAGUACUUUUUAAAGUGUAGCUAGCGUAGAUGGAACAGAGGCAGGGUGAUGCCAGUGGUGGUGAUGGUGGCUACCUGACCAGGUGGCGUCGAUCUACCG ..((.(((((.(((((....)))))))))).))((((((.((.((..((((......))))..))............(((((((....))))))).)).)))))).. ( -39.40) >DroEre_CAF1 41382 107 - 1 UUACUACACAGCAUUUUUUAGAGUGUAGCUAGGGUAGAUGGAGCAGAGGCGGGCUGAUGGCGGUGGUGGUGAUGGUGGCUACCUGACCAGGUGGCGUCGAUCUACCG ...((((((.............))))))....(((((((.((((....))..((((.((((((((((.(......).)))))).).)))..)))).)).))))))). ( -35.12) >consensus UUACAAGCCAGCACUUUUUAAAGUGUAGCUAGCGUAGAUGGAACAGAGGCAGGGUGAUGCCAGUGGUGGUGAUGGUGGCUACCUGACCAGGUGGCGUCGAUCUACCG ..((((((((.(((((....)))))))))))))((((((.((.....((((......))))................(((((((....))))))).)).)))))).. (-29.85 = -31.97 + 2.12)

| Location | 10,326,967 – 10,327,078 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 83.93 |
| Mean single sequence MFE | -24.11 |
| Consensus MFE | -14.02 |
| Energy contribution | -17.28 |
| Covariance contribution | 3.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
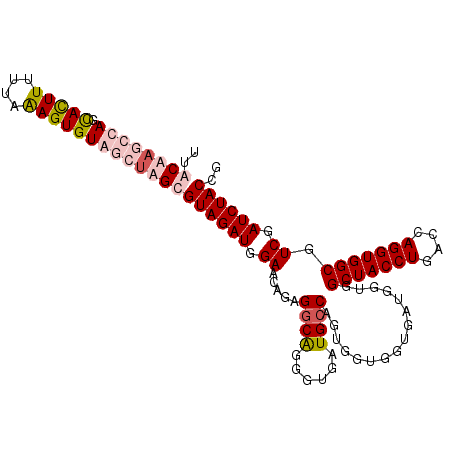
>2R_DroMel_CAF1 10326967 111 + 20766785 CACCACCACUGGCAUCACCCUGCCUCUGUUCCAUCUACGCUAGAUACACUUUAAAAAAUGUUGGUAAGUAAGAUGGGAUCAUUUAAUCAGCAUUCUAUUCUUUUGCUUUUA ......((..((((......))))..))((((((((((((((...(((.((....)).)))))))..)).))))))))..........((((...........)))).... ( -17.90) >DroSec_CAF1 41428 111 + 1 CACCACCAUUGGCAUCACCCUGCCUCGGUUCCAUCUACGCUAGCUACACUUUAAAAAGUACUAGCUUGCUAGAUGGGAUCAUUUAAUCAUCAUUCUAUUCUUUUGCUUUCG ..........((((......))))..(((((((((((.((.(((((.((((....))))..))))).)))))))))))))............................... ( -30.60) >DroSim_CAF1 41620 111 + 1 CACCACCACUGGCAUCACCCUGCCUCUGUUCCAUCUACGCUAGCUACACUUUAAAAAGUACUAGCUUGCAAGAUGGGAUCAUUAAAUCAUCAUUCUAUUCUUUUGCUUUCG ......((..((((......))))..))((((((((..((.(((((.((((....))))..))))).)).))))))))................................. ( -24.30) >DroEre_CAF1 41418 111 + 1 CACCACCACCGCCAUCAGCCCGCCUCUGCUCCAUCUACCCUAGCUACACUCUAAAAAAUGCUGUGUAGUAAGAUGGGAUCAUUCACUGAUCUUUAUAUCGUUGUGAUUUAG ...(((.((..((((((((........)))............(((((((.............)))))))..)))))(((((.....)))))........)).)))...... ( -23.62) >consensus CACCACCACUGGCAUCACCCUGCCUCUGUUCCAUCUACGCUAGCUACACUUUAAAAAAUACUAGCUAGCAAGAUGGGAUCAUUUAAUCAUCAUUCUAUUCUUUUGCUUUCG ..........((((......))))...(((((((((..(((((((((((((....))))).)))))))).)))))))))................................ (-14.02 = -17.28 + 3.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:50 2006