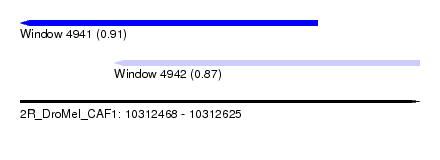
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,312,468 – 10,312,625 |
| Length | 157 |
| Max. P | 0.913113 |

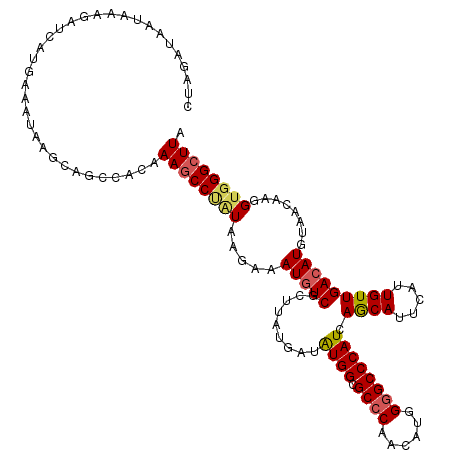
| Location | 10,312,468 – 10,312,585 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 91.97 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -26.14 |
| Energy contribution | -26.22 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10312468 117 - 20766785 CUAAGAAAUAUCGCCUAUGAUAUGGCGCCCAACAUGGGGCCCAUCAGCAUUCAUUGUUGACAUGUAACAAGGUGGGCUUAACGGGAAACCCCCUUAAUCUCUGCUAAUCUCUCCCUA .((((.......((((((....(((.((((......)))))))((((((.....))))))...........)))))).....((....))..))))..................... ( -31.20) >DroSec_CAF1 26884 117 - 1 GUAAGAAAUGUCGCUUAUAAUAUGGCGCCCAACAUGGGGCCCAUCAGCAUUCAUUGUUGACAUGUAACAAGGUUGGCUUAACGGGAAACCCCCUUAAUCUCUGCUAAUCUCUCCCCA (..(((..(((..(........(((.((((......)))))))((((((.....))))))...)..))).(((((((((((.(((....))).)))).....))))))))))..).. ( -32.20) >DroSim_CAF1 27199 117 - 1 AUAAGAAAUGUCGUUUAUGAUGUGGCGCCCAACAUGGGGCCCAUCAGCAUUCAUUGUUGACAUGUAACAAGGUGGGCUUAACGGGAAACCCCCUUAAUCUCUGCUAAUCUCUCCCCA ...(((.(((((......((((.(((.(((.....))))))))))((((.....))))))))).......((..((.((((.(((....))).)))).).)..))..)))....... ( -32.70) >DroEre_CAF1 26988 117 - 1 AUAAGAAAUGUCGCUUACUAUAUGGCGCCCAACGAGGGGCCCAUCAGCAUUCAUUGUUGACAUGUAACAAGGUGGGCUUAACGGGAAACCCACUUAAUCUCUACUAAUCUCUCCCCU .((((.......(((((((...(((.((((......)))))))((((((.....))))))..........))))))).....((....))..))))..................... ( -31.60) >DroYak_CAF1 27680 116 - 1 AUAGGAAAUGUCGCUUAUGAUAUGGCGCCCAACGAGG-GCCCAUCAACAUUUACUGUUGACAUGUAACAAGGUGGGCUUAACGGGAAAGCCCCUUAAUCUCUACUAAUCUCUCCCCU ...(((..(((..(..(((...(((.((((.....))-)))))((((((.....))))))))))..))).((((((.((((.(((....))).)))).).)))))......)))... ( -32.30) >consensus AUAAGAAAUGUCGCUUAUGAUAUGGCGCCCAACAUGGGGCCCAUCAGCAUUCAUUGUUGACAUGUAACAAGGUGGGCUUAACGGGAAACCCCCUUAAUCUCUGCUAAUCUCUCCCCA ...(((..(((..(........(((.((((......)))))))((((((.....))))))...)..))).((((((.((((.(((....))).)))).).)))))..)))....... (-26.14 = -26.22 + 0.08)

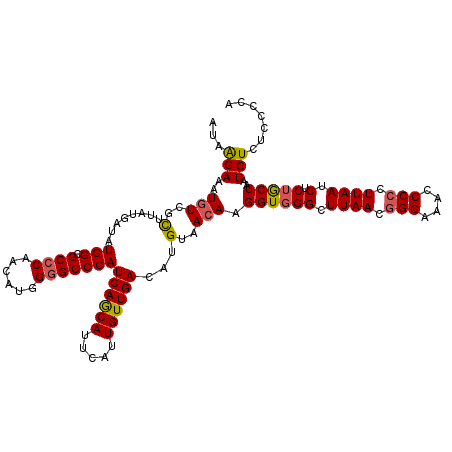
| Location | 10,312,505 – 10,312,625 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.83 |
| Mean single sequence MFE | -31.59 |
| Consensus MFE | -24.60 |
| Energy contribution | -25.00 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10312505 120 - 20766785 CUAGAUAAUACAGAUCAUAAAAUAAGCAGACACAAAACCUCUAAGAAAUAUCGCCUAUGAUAUGGCGCCCAACAUGGGGCCCAUCAGCAUUCAUUGUUGACAUGUAACAAGGUGGGCUUA ...(((.......)))......((((((((.........)))........((((((.((...(((.((((......)))))))((((((.....)))))).......))))))))))))) ( -27.90) >DroSec_CAF1 26921 120 - 1 CUAGAUAGUAAAGAUCAUGAAAUAAGCAGCCACAAAGCCUGUAAGAAAUGUCGCUUAUAAUAUGGCGCCCAACAUGGGGCCCAUCAGCAUUCAUUGUUGACAUGUAACAAGGUUGGCUUA ...(((.......)))...........(((((....((((.......(((((.........((((.((((......)))))))).((((.....)))))))))......))))))))).. ( -29.62) >DroSim_CAF1 27236 120 - 1 CUAGAUAAUAAAGAUCAAGCAAUAAGCAGCCACAAAGCCUAUAAGAAAUGUCGUUUAUGAUGUGGCGCCCAACAUGGGGCCCAUCAGCAUUCAUUGUUGACAUGUAACAAGGUGGGCUUA ...(((.......)))..((.....)).......((((((((.....(((((......((((.(((.(((.....))))))))))((((.....)))))))))........)))))))). ( -34.42) >DroEre_CAF1 27025 120 - 1 CUAGACAAUUUAGAUCAUGAAAUAAGCAGCCACAAAGCCCAUAAGAAAUGUCGCUUACUAUAUGGCGCCCAACGAGGGGCCCAUCAGCAUUCAUUGUUGACAUGUAACAAGGUGGGCUUA (((((...))))).....................((((((((......(((..(........(((.((((......)))))))((((((.....))))))...)..)))..)))))))). ( -31.80) >DroYak_CAF1 27717 119 - 1 CUAGAUAAAUUAGGCCAUGAAAUAAGCAGCCACUAAGCCCAUAGGAAAUGUCGCUUAUGAUAUGGCGCCCAACGAGG-GCCCAUCAACAUUUACUGUUGACAUGUAACAAGGUGGGCUUA ............(((.............)))..(((((((((......(((..(..(((...(((.((((.....))-)))))((((((.....))))))))))..)))..))))))))) ( -34.22) >consensus CUAGAUAAUAAAGAUCAUGAAAUAAGCAGCCACAAAGCCUAUAAGAAAUGUCGCUUAUGAUAUGGCGCCCAACAUGGGGCCCAUCAGCAUUCAUUGUUGACAUGUAACAAGGUGGGCUUA ..................................((((((((.....(((((.........((((.((((......)))))))).((((.....)))))))))........)))))))). (-24.60 = -25.00 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:45 2006