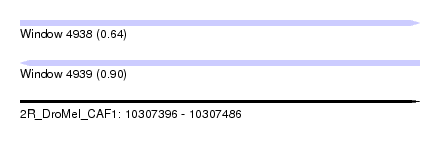
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,307,396 – 10,307,486 |
| Length | 90 |
| Max. P | 0.896431 |

| Location | 10,307,396 – 10,307,486 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 78.42 |
| Mean single sequence MFE | -24.15 |
| Consensus MFE | -14.57 |
| Energy contribution | -14.94 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10307396 90 + 20766785 UGCGCUCUCGUUUUCUGAU-----GCGAACGGGCUAAAUGGGCAAGGAACGCCCACAAAAACGAAGAGAAAU--------CGAGAAAGUAGUGCCACCGAUUG .(((((((((.(((((...-----((......))....(((((.......)))))...........))))).--------))))).....))))......... ( -23.00) >DroSec_CAF1 21861 93 + 1 UGCGCGCUCGUUUUCUGAU-----GCGAACGGGCUAAAUGGGCAGGGAACGCCCACAAAAACAGAGAGAAAUCGAGAAAUCGAGAAAGUCCGGCAAUC----- (((.((((((.....))).-----)))..((((((...(((((.(....))))))................((((....))))...)))))))))...----- ( -30.90) >DroSim_CAF1 21834 85 + 1 UGCGCGCUCGUUUUCUGAU-----GCGAACGGGCUAAAUGGGCAGGGAACGCCCACAAAAACGGAGAGAAAU--------CGAGAAAGUCGGGCAAUC----- (((.(((((((((.(....-----).))))))))....(((((.(....))))))..........((....)--------).........).)))...----- ( -26.20) >DroEre_CAF1 21971 82 + 1 UGCGCGCUCGUU-UCUGAU-----GCGAACGGGCUAAAUGGGCAAGAAACGCCCACAAAAGCGGAGGGAAAU--------CGAGAAAAGCGGGAAA------- ..(((.((((((-(((..(-----(((......)....(((((.......))))).....)))...))))).--------))))....))).....------- ( -22.70) >DroYak_CAF1 22330 87 + 1 UGCGCGCUCGUU-UCUGAUGCGAUGCGAACGGGCUAAAUGGGCAAGAAACGCCCACAAAAGCGGAGAGAAAC--------UGAGAAAAGCUGGAAA------- .((...((((((-(((..(((...((......))....(((((.......))))).....)))...))))))--------.)))....))......------- ( -24.00) >DroAna_CAF1 25335 73 + 1 UGCGAGCUCGUUUUCUGAU-----CCGAAUGGGCUAAAUGGGCAAGAAACGCCCACAAA-ACGGAGAAAAAU--------UGAGAGG---------------- ..(((((((((((......-----..)))))))))...(((((.......)))))....-.)).........--------.......---------------- ( -18.10) >consensus UGCGCGCUCGUUUUCUGAU_____GCGAACGGGCUAAAUGGGCAAGAAACGCCCACAAAAACGGAGAGAAAU________CGAGAAAGUCGGGCAA_______ .....((((((((.............))))))))....(((((.......)))))................................................ (-14.57 = -14.94 + 0.36)

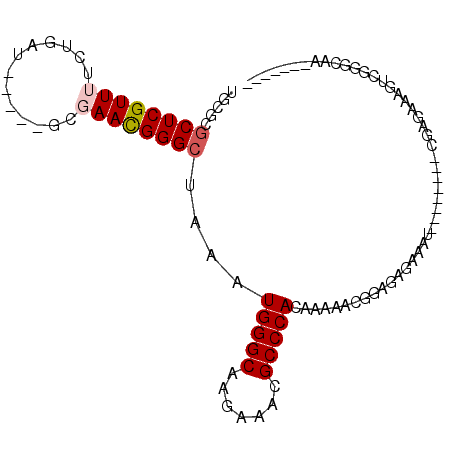
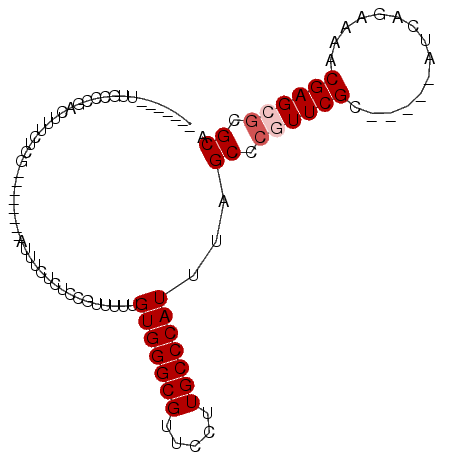
| Location | 10,307,396 – 10,307,486 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 78.42 |
| Mean single sequence MFE | -22.40 |
| Consensus MFE | -17.18 |
| Energy contribution | -17.68 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10307396 90 - 20766785 CAAUCGGUGGCACUACUUUCUCG--------AUUUCUCUUCGUUUUUGUGGGCGUUCCUUGCCCAUUUAGCCCGUUCGC-----AUCAGAAAACGAGAGCGCA .((((((.((........)))))--------))).((((.((((((((((((((.....)))))))...((......))-----....))))))))))).... ( -23.80) >DroSec_CAF1 21861 93 - 1 -----GAUUGCCGGACUUUCUCGAUUUCUCGAUUUCUCUCUGUUUUUGUGGGCGUUCCCUGCCCAUUUAGCCCGUUCGC-----AUCAGAAAACGAGCGCGCA -----......((((.....((((....))))......)))).....(((((((.....)))))))...((.((((((.-----.........)))))).)). ( -23.30) >DroSim_CAF1 21834 85 - 1 -----GAUUGCCCGACUUUCUCG--------AUUUCUCUCCGUUUUUGUGGGCGUUCCCUGCCCAUUUAGCCCGUUCGC-----AUCAGAAAACGAGCGCGCA -----...(((.((.....((((--------.(((((..........(((((((.....)))))))...((......))-----...))))).)))))).))) ( -20.70) >DroEre_CAF1 21971 82 - 1 -------UUUCCCGCUUUUCUCG--------AUUUCCCUCCGCUUUUGUGGGCGUUUCUUGCCCAUUUAGCCCGUUCGC-----AUCAGA-AACGAGCGCGCA -------.....(((((((((.(--------((........(((...(((((((.....)))))))..)))........-----))))))-)..))))).... ( -23.19) >DroYak_CAF1 22330 87 - 1 -------UUUCCAGCUUUUCUCA--------GUUUCUCUCCGCUUUUGUGGGCGUUUCUUGCCCAUUUAGCCCGUUCGCAUCGCAUCAGA-AACGAGCGCGCA -------......((........--------..........(((...(((((((.....)))))))..))).((((((..((......))-..)))))).)). ( -25.20) >DroAna_CAF1 25335 73 - 1 ----------------CCUCUCA--------AUUUUUCUCCGU-UUUGUGGGCGUUUCUUGCCCAUUUAGCCCAUUCGG-----AUCAGAAAACGAGCUCGCA ----------------...(((.--------..((((((..((-(..(((((((.....)))))))..)))((....))-----...)))))).)))...... ( -18.20) >consensus _______UUGCCCGACUUUCUCG________AUUUCUCUCCGUUUUUGUGGGCGUUCCUUGCCCAUUUAGCCCGUUCGC_____AUCAGAAAACGAGCGCGCA ...............................................(((((((.....)))))))...((.((((((...............)))))).)). (-17.18 = -17.68 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:42 2006