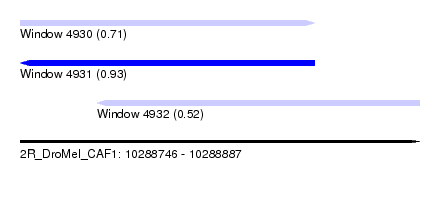
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,288,746 – 10,288,887 |
| Length | 141 |
| Max. P | 0.930234 |

| Location | 10,288,746 – 10,288,850 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.70 |
| Mean single sequence MFE | -34.92 |
| Consensus MFE | -21.67 |
| Energy contribution | -22.65 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
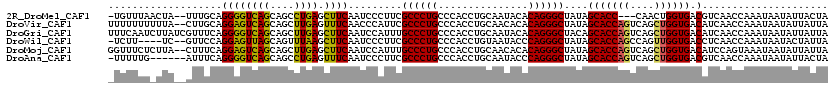
>2R_DroMel_CAF1 10288746 104 + 20766785 UUGGGGCCU--GCCAAU-----------AGCCUCAUUUAGUAGUAAUAUUAUUUGGUUGACGUCACCAGUUG---GGUGCUAUAGCCCUGUGUAUUGCAGGUGGGCAGGGCGAAGGGAUU ....(.(((--(((.((-----------(((((((..(((((....))))).(((((.(....)))))).))---)).)))))..((((((.....))))).))))))).)......... ( -35.70) >DroVir_CAF1 8903 109 + 1 UCGGAGCCUGGGCCAAC-----------AGCUUCCCUUAAUAAUAAUAUUAUUUGGUUGAUGUCACCAGCUGACUGGUGCUAUAGCCCUGUGUGUUGCAGGUGGGCAGGGCGAAUGGGUU (((..((((..(((..(-----------((((.....(((((....)))))...)))))(((.((((((....)))))).)))..((((((.....))))).)))).))))...)))... ( -33.30) >DroGri_CAF1 4912 109 + 1 CAAGAGCAGUGGCCAAU-----------AGCCUCAUUUAAUAAUAAUAUUAUUUGGUUGAUGUCACCAGCUGACUGGUGCUGUAGCCCUGUGUAUUGCAGGUGGGCAGGGCAAAUGGAUU .....((((((((...(-----------((((.....(((((....)))))...)))))..)))(((((....)))))))))).(((((((.(((.....))).)))))))......... ( -34.40) >DroWil_CAF1 22355 115 + 1 ---GAGCCC--UCCAAUUUAUUUCUUUUUUUUUCGUUUAAUAAUAGUAUUAUUUGGUUGAGGUCACCAACUGGCUGGUGCUAUAGCCCUGGGUAUUACAGGUGGGCAGGGCGAAGGGAUU ---...(((--(.(............................(((((((((.(..((((.(....)))))..).))))))))).((((((..(((.....)))..))))))).))))... ( -33.40) >DroMoj_CAF1 5308 107 + 1 UC-CAGCCA-GGACAUC-----------CGACUAACUUAAUAAUAAUAUUAUUUACUGGAUGUCACCAGCUGACUGGUGCUAUAGCCCUGUGUGUUGCAGGUGGGCAGGGCAAAUGGAUU ((-((((..-.((((((-----------((..(((..(((((....))))).))).))))))))(((((....))))))))...(((((((.(.........).)))))))....))).. ( -35.70) >DroAna_CAF1 7502 107 + 1 UGGUGGCCU--GCCAAU-----------AGCCUCAUUUAGUAGUAAUAUUAUUUGGUUGACGUCACCAGCUGACUGGUGCUAUAGCCCUGGGUAUUGCAGGUGGGCAGGGCGAAGGGAUU ...((.(((--(((...-----------.((((.........((((((((....(((((..(.((((((....)))))))..)))))...)))))))))))).)))))).))........ ( -37.00) >consensus UCGGAGCCU__GCCAAU___________AGCCUCAUUUAAUAAUAAUAUUAUUUGGUUGAUGUCACCAGCUGACUGGUGCUAUAGCCCUGUGUAUUGCAGGUGGGCAGGGCGAAGGGAUU ......(((...................((((.....(((((....)))))...))))...(.((((((....)))))))....(((((((.(((.....))).)))))))...)))... (-21.67 = -22.65 + 0.98)

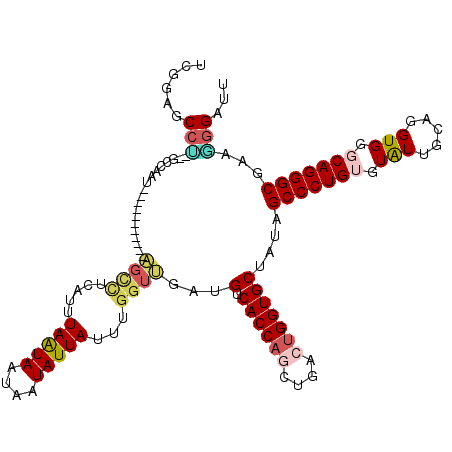
| Location | 10,288,746 – 10,288,850 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.70 |
| Mean single sequence MFE | -28.10 |
| Consensus MFE | -18.04 |
| Energy contribution | -18.77 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10288746 104 - 20766785 AAUCCCUUCGCCCUGCCCACCUGCAAUACACAGGGCUAUAGCACC---CAACUGGUGACGUCAACCAAAUAAUAUUACUACUAAAUGAGGCU-----------AUUGGC--AGGCCCCAA ...........((((((..((((.......))))...(((((...---....((((.......)))).............((.....)))))-----------)).)))--)))...... ( -22.80) >DroVir_CAF1 8903 109 - 1 AACCCAUUCGCCCUGCCCACCUGCAACACACAGGGCUAUAGCACCAGUCAGCUGGUGACAUCAACCAAAUAAUAUUAUUAUUAAGGGAAGCU-----------GUUGGCCCAGGCUCCGA .........(((.(((......))).......(((((((((((((((....)))))........((...(((((....)))))..))..)))-----------).)))))).)))..... ( -32.20) >DroGri_CAF1 4912 109 - 1 AAUCCAUUUGCCCUGCCCACCUGCAAUACACAGGGCUACAGCACCAGUCAGCUGGUGACAUCAACCAAAUAAUAUUAUUAUUAAAUGAGGCU-----------AUUGGCCACUGCUCUUG ....(((((((((((...............))))))....(((((((....)))))).).......................))))).(((.-----------....))).......... ( -26.46) >DroWil_CAF1 22355 115 - 1 AAUCCCUUCGCCCUGCCCACCUGUAAUACCCAGGGCUAUAGCACCAGCCAGUUGGUGACCUCAACCAAAUAAUACUAUUAUUAAACGAAAAAAAAAGAAAUAAAUUGGA--GGGCUC--- ...((((((((((((...............)))))).....((((((....)))))).................................................)))--)))...--- ( -26.06) >DroMoj_CAF1 5308 107 - 1 AAUCCAUUUGCCCUGCCCACCUGCAACACACAGGGCUAUAGCACCAGUCAGCUGGUGACAUCCAGUAAAUAAUAUUAUUAUUAAGUUAGUCG-----------GAUGUCC-UGGCUG-GA .........((((((...............))))))...........(((((..(.((((((((.(((.(((((....)))))..))).).)-----------)))))))-..))))-). ( -34.26) >DroAna_CAF1 7502 107 - 1 AAUCCCUUCGCCCUGCCCACCUGCAAUACCCAGGGCUAUAGCACCAGUCAGCUGGUGACGUCAACCAAAUAAUAUUACUACUAAAUGAGGCU-----------AUUGGC--AGGCCACCA .........(.((((((..((((.......))))(((...(((((((....)))))).).............((((.......)))).))).-----------...)))--))).).... ( -26.80) >consensus AAUCCAUUCGCCCUGCCCACCUGCAAUACACAGGGCUAUAGCACCAGUCAGCUGGUGACAUCAACCAAAUAAUAUUAUUAUUAAAUGAGGCU___________AUUGGC__AGGCUCCGA .........(((..(((..((((.......))))......(((((((....)))))).)...............................................)))...)))..... (-18.04 = -18.77 + 0.72)

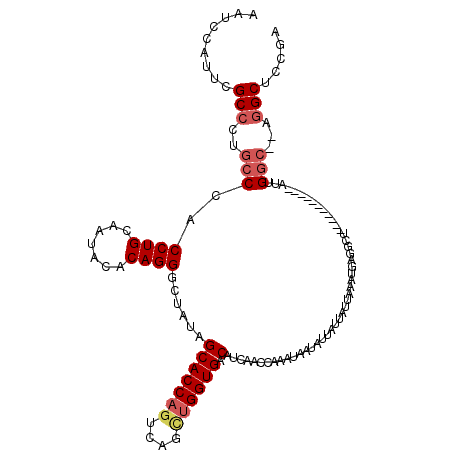

| Location | 10,288,773 – 10,288,887 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.83 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -22.69 |
| Energy contribution | -21.94 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.515804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
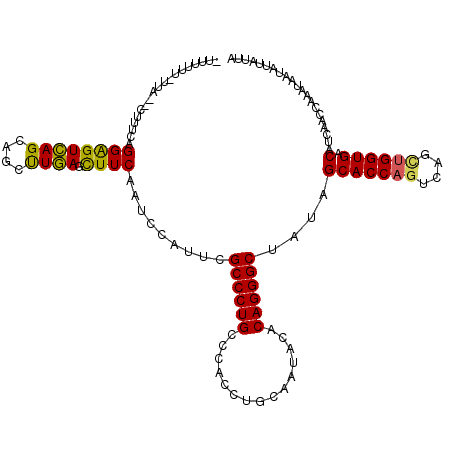
>2R_DroMel_CAF1 10288773 114 - 20766785 -UGUUUAACUA--UUUGCAGGGGUCAGCAGCCUGAGCUUCAAUCCCUUCGCCCUGCCCACCUGCAAUACACAGGGCUAUAGCACC---CAACUGGUGACGUCAACCAAAUAAUAUUACUA -(((((.....--.(((((((((...((((..((((..........))))..)))))).)))))))......((.......((((---.....)))).......)))))))......... ( -25.64) >DroVir_CAF1 8932 118 - 1 UUUUUUUUUUA--CUUGCAGGAGUCAGCAGCUUGAGUUUCAACCCAUUCGCCCUGCCCACCUGCAACACACAGGGCUAUAGCACCAGUCAGCUGGUGACAUCAACCAAAUAAUAUUAUUA ...........--.(((((((.(...((((..(((((........)))))..)))).).)))))))......((.......((((((....)))))).......)).............. ( -30.14) >DroGri_CAF1 4941 120 - 1 UUUCAAUCUUAUCGUUUCAGGGGUCAGCAGCUUGAGCUUCAAUCCAUUUGCCCUGCCCACCUGCAAUACACAGGGCUACAGCACCAGUCAGCUGGUGACAUCAACCAAAUAAUAUUAUUA ....(((..(((..(((..((.((((.(((((.((.((...........((((((...............)))))).........))))))))).)))).....)))))..)))..))). ( -26.31) >DroWil_CAF1 22390 113 - 1 -UCUU----UC--GUUCCAGGAGUUAGCAGUUUAAGCUUCAAUCCCUUCGCCCUGCCCACCUGUAAUACCCAGGGCUAUAGCACCAGCCAGUUGGUGACCUCAACCAAAUAAUACUAUUA -....----..--......(((...(((.......)))....)))....((((((...............)))))).....((((((....))))))....................... ( -22.46) >DroMoj_CAF1 5335 118 - 1 GGUUUCUCUUA--CUUUCAGGAGUCAGCAGCUUGAGCUUCAAUCCAUUUGCCCUGCCCACCUGCAACACACAGGGCUAUAGCACCAGUCAGCUGGUGACAUCCAGUAAAUAAUAUUAUUA ........(((--((....(((((((.(((((.((.((...........((((((...............)))))).........))))))))).)))).))))))))............ ( -29.51) >DroAna_CAF1 7529 113 - 1 -UUUUUG------AUUUCAGGGGUCAGCAGCCUGAGUUUCAAUCCCUUCGCCCUGCCCACCUGCAAUACCCAGGGCUAUAGCACCAGUCAGCUGGUGACGUCAACCAAAUAAUAUUACUA -...(((------(.((((((.(....)..))))))..)))).......((((((...............))))))....(((((((....)))))).)..................... ( -28.66) >consensus _UUUUUU_UUA__CUUUCAGGAGUCAGCAGCUUGAGCUUCAAUCCAUUCGCCCUGCCCACCUGCAAUACACAGGGCUAUAGCACCAGUCAGCUGGUGACAUCAACCAAAUAAUAUUAUUA ...................((((((((....)))).)))).........((((((...............))))))....(((((((....)))))).)..................... (-22.69 = -21.94 + -0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:36 2006